Analysis of an Autographa californica nucleopolyhedrovirus lef-11 knockout: LEF-11 is essential for viral DNA replication
- PMID: 11861844
- PMCID: PMC135986
- DOI: 10.1128/jvi.76.6.2770-2779.2002
Analysis of an Autographa californica nucleopolyhedrovirus lef-11 knockout: LEF-11 is essential for viral DNA replication
Abstract
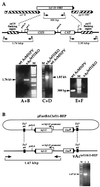
The Autographa californica nucleopolyhedrovirus (AcMNPV) lef-11 gene was previously identified by transient late expression assays as a gene important for viral late gene expression. The lef-11 gene was not previously identified as necessary for DNA replication in transient origin-dependent plasmid DNA replication assays. To examine the role of lef-11 in the context of the infection cycle, we generated a deletion of the lef-11 gene by recombination in an AcMNPV genome propagated as a BACmid in Escherichia coli. The resulting AcMNPV lef-11-null BACmid (vAc(lef11KO)) was unable to propagate in cell culture, although a "repair" AcMNPV BACmid (vAc(lef11KO-REP)), which was generated by transposition of the lef-11 gene into the polyhedrin locus of the vAc(lef11KO) BACmid, was able to replicate in a manner similar to wild-type or control AcMNPV viruses. Thus, the lef-11 gene is essential for viral replication in Sf9 cells. The vAc(lef11KO) BACmid was examined to determine if the defect in viral replication resulted from a defect in DNA replication or from a defect in late transcription. The lef-11-null BACmid and control BACmids were transfected into Sf9 cells, and viral DNA replication was monitored. The viral DNA genome of the lef-11-null BACmid (vAc(lef11KO)) was not amplified, whereas replication and amplification of the genomes of the repair BACmid (vAc(lef11KO-REP)), wild-type AcMNPV, and a nonpropagating gp64-null control BACmid (vAc(GUSgp64KO)) were readily detected. Northern blot analysis of transcripts from selected early, late, and very late genes showed that late and very late transcription was absent in cells transfected with the lef-11-null BACmid. Thus, in contrast to prior studies using transient replication and late expression assays, studies of a lef-11-null BACmid indicate that LEF-11 is required for viral DNA replication during the infection cycle.
Figures





Similar articles
-
Analysis of an Autographa californica multicapsid nucleopolyhedrovirus lef-6-null virus: LEF-6 is not essential for viral replication but appears to accelerate late gene transcription.J Virol. 2002 Jun;76(11):5503-14. doi: 10.1128/jvi.76.11.5503-5514.2002. J Virol. 2002. PMID: 11991978 Free PMC article.
-
Characterization of a baculovirus lacking the alkaline nuclease gene.J Virol. 2004 Oct;78(19):10650-6. doi: 10.1128/JVI.78.19.10650-10656.2004. J Virol. 2004. PMID: 15367632 Free PMC article.
-
The Autographa californica multiple nucleopolyhedrovirus lef-5 gene is required for productive infection.Virology. 2011 Jul 20;416(1-2):54-64. doi: 10.1016/j.virol.2011.04.019. Epub 2011 May 23. Virology. 2011. PMID: 21601232
-
Autographa californica multiple nucleopolyhedrovirus nucleocapsid assembly is interrupted upon deletion of the 38K gene.J Virol. 2006 Dec;80(23):11475-85. doi: 10.1128/JVI.01155-06. Epub 2006 Sep 20. J Virol. 2006. PMID: 16987976 Free PMC article.
-
Baculovirus Molecular Biology [Internet].3rd edition. Bethesda (MD): National Center for Biotechnology Information (US); 2013. 3rd edition. Bethesda (MD): National Center for Biotechnology Information (US); 2013. PMID: 24479205 Free Books & Documents. Review.
Cited by
-
Baculovirus DNA replication-specific expression factors trigger apoptosis and shutoff of host protein synthesis during infection.J Virol. 2009 Nov;83(21):11123-32. doi: 10.1128/JVI.01199-09. Epub 2009 Aug 12. J Virol. 2009. PMID: 19706708 Free PMC article.
-
Identification of a domain of the baculovirus Autographa californica multiple nucleopolyhedrovirus single-strand DNA-binding protein LEF-3 essential for viral DNA replication.J Virol. 2010 Jun;84(12):6153-62. doi: 10.1128/JVI.00115-10. Epub 2010 Mar 31. J Virol. 2010. PMID: 20357098 Free PMC article.
-
A sensitive assay for scrutiny of Autographa californica multiple nucleopolyhedrovirus genes using CRISPR-Cas9.Appl Microbiol Biotechnol. 2023 Jul;107(13):4323-4335. doi: 10.1007/s00253-023-12462-y. Epub 2023 May 26. Appl Microbiol Biotechnol. 2023. PMID: 37233755
-
Functional analysis of the transmembrane (TM) domain of the Autographa californica multicapsid nucleopolyhedrovirus GP64 protein: substitution of heterologous TM domains.J Virol. 2008 Apr;82(7):3329-41. doi: 10.1128/JVI.02104-07. Epub 2008 Jan 23. J Virol. 2008. PMID: 18216100 Free PMC article.
-
IE1 of Autographa californica Multiple Nucleopolyhedrovirus Activates Low Levels of Late Gene Expression in the Absence of Virus RNA Polymerase.Microbiol Spectr. 2023 Feb 14;11(1):e0343222. doi: 10.1128/spectrum.03432-22. Epub 2022 Dec 13. Microbiol Spectr. 2023. PMID: 36511657 Free PMC article.
References
-
- Ahrens, C. H., D. J. Leisy, and G. F. Rohrmann. 1996. Baculovirus DNA replication, p. 855-872. In M. L. DePamphilis (ed.), DNA replication in eukaryotic cells. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
-
- Ayres, M. D., S. C. Howard, J. Kuzio, M. Lopez-Ferber, and R. D. Possee. 1994. The complete DNA sequence of Autographa californica nuclear polyhedrosis virus. Virology 202:586-605. - PubMed
-
- Bideshi, D. K., and B. A. Federici. 2000. The Trichoplusia ni granulovirus helicase is unable to support replication of Autographa californica multicapsid nucleopolyhedrovirus in cells and larvae of T. ni. J. Gen. Virol. 81:1593-1599. - PubMed
-
- Blissard, G., B. Black, N. Crook, B. A. Keddie, R. Possee, G. Rohrmann, D. Theilmann, and L. E. Volkman. 2000. Baculoviridae: taxonomic structure and properties of the family, p. 195-202. In M.H.V. van Regenmortel et al. (ed.), Virus taxonomy. The classification and nomenclature of viruses. The seventh report of the International Committee for the Taxonomy of Viruses. Academic Press, San Diego, Calif.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

