Selectable subgenomic and genome-length dicistronic RNAs derived from an infectious molecular clone of the HCV-N strain of hepatitis C virus replicate efficiently in cultured Huh7 cells
- PMID: 11861865
- PMCID: PMC135991
- DOI: 10.1128/jvi.76.6.2997-3006.2002
Selectable subgenomic and genome-length dicistronic RNAs derived from an infectious molecular clone of the HCV-N strain of hepatitis C virus replicate efficiently in cultured Huh7 cells
Abstract
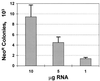
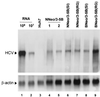
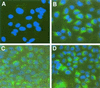
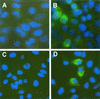
Dicistronic, selectable subgenomic replicons derived from the Con1 strain of hepatitis C virus (HCV) are capable of autonomous replication in cultured Huh7 cells (Lohmann et al., Science 285:110-113, 1999). However, adaptive mutations in the NS3, NS5A, and/or NS5B proteins are required for efficient replication of these RNAs and increase by orders of magnitude the numbers of G418-resistant colonies selected following transfection of Huh7 cells. Here, we demonstrate that a subgenomic replicon (NNeo/3-5B) derived from an infectious molecular clone of a second genotype 1b virus, HCV-N (Beard et al., Hepatology 30:316-324, 1999) is also capable of efficient replication in Huh7 cells. G418-resistant cells selected following transfection with NNeo/3-5B RNA contained abundant NS5A antigen and HCV RNA detectable by Northern analysis. Replicon RNA in one of three clonally isolated cell lines contained no mutations in the NS3-NS5B polyprotein, confirming that adaptive mutations are not required for efficient replication in these cells. However, the deletion of a unique 4-amino-acid insertion that is present within the interferon sensitivity-determining region (ISDR) of the NS5A protein in wild-type HCV-N drastically decreased the number of G418-resistant colonies obtained following transfection of Huh7 cells. This effect could be reversed by inclusion of a previously described Con1 cell culture-adaptive mutation (S2005-->I), confirming that this natural insertion has a controlling role in determining the replication capacity of wild-type HCV-N RNA in Huh7 cells. Additional selectable, dicistronic RNAs encoding NS2-NS5B, E1-NS5B, or the full-length HCV polyprotein were also capable of replication and gave rise to G418-resistant cell clones following transfection of Huh7 cells. We conclude that RNA derived from this documented infectious molecular clone has a unique capacity for replication in Huh7 cells in the absence of additional cell culture-adaptive mutations.
Figures







Similar articles
-
Introduction of NS5A mutations enables subgenomic HCV replicon derived from chimpanzee-infectious HC-J4 isolate to replicate efficiently in Huh-7 cells.J Viral Hepat. 2004 Sep;11(5):394-403. doi: 10.1111/j.1365-2893.2004.00525.x. J Viral Hepat. 2004. PMID: 15357644
-
Dominant negative effect of wild-type NS5A on NS5A-adapted subgenomic hepatitis C virus RNA replicon.J Gen Virol. 2004 Jul;85(Pt 7):1867-1875. doi: 10.1099/vir.0.80006-0. J Gen Virol. 2004. PMID: 15218171
-
Replication studies using genotype 1a subgenomic hepatitis C virus replicons.J Virol. 2003 May;77(9):5352-9. doi: 10.1128/jvi.77.9.5352-5359.2003. J Virol. 2003. PMID: 12692237 Free PMC article.
-
[Interferon resistance and ISDR (interferon sensitivity determining region)].Nihon Rinsho. 2006 Jul;64(7):1249-53. Nihon Rinsho. 2006. PMID: 16838640 Review. Japanese.
-
Recent advances in the molecular biology of hepatitis C virus.J Mol Biol. 2001 Oct 26;313(3):451-64. doi: 10.1006/jmbi.2001.5055. J Mol Biol. 2001. PMID: 11676530 Review.
Cited by
-
Identification of residues in the hepatitis C virus core protein that are critical for capsid assembly in a cell-free system.J Virol. 2005 Jun;79(11):6814-26. doi: 10.1128/JVI.79.11.6814-6826.2005. J Virol. 2005. PMID: 15890921 Free PMC article.
-
Replication of hepatitis C virus (HCV) RNA in mouse embryonic fibroblasts: protein kinase R (PKR)-dependent and PKR-independent mechanisms for controlling HCV RNA replication and mediating interferon activities.J Virol. 2006 Aug;80(15):7364-74. doi: 10.1128/JVI.00586-06. J Virol. 2006. PMID: 16840317 Free PMC article.
-
Interactions between Hsp90 and oncogenic viruses: implications for viral cancer therapeutics.Am J Cancer Res. 2011;1(6):763-72. Epub 2011 Jun 5. Am J Cancer Res. 2011. PMID: 22016826 Free PMC article.
-
Establishment of infectious HCV virion-producing cells with newly designed full-genome replicon RNA.Arch Virol. 2011 Feb;156(2):295-304. doi: 10.1007/s00705-010-0859-x. Epub 2011 Jan 19. Arch Virol. 2011. PMID: 21246385 Free PMC article.
-
Development of intergenotypic chimeric replicons to determine the broad-spectrum antiviral activities of hepatitis C virus polymerase inhibitors.Antimicrob Agents Chemother. 2008 Oct;52(10):3523-31. doi: 10.1128/AAC.00533-08. Epub 2008 Aug 11. Antimicrob Agents Chemother. 2008. PMID: 18694956 Free PMC article.
References
-
- Alter, M. J., H. S. Margolis, K. Krawczynski, F. N. Judson, A. Mares, W. J. Alexander, P. Y. Hu, J. K. Miller, M. A. Gerber, and R. E. Sampliner. 1992. The natural history of community-acquired hepatitis C in the United States. The Sentinel Counties Chronic non-A, non-B Hepatitis Study Team. N. Engl. J. Med. 327:1899-1905. - PubMed
-
- Beard, M. R., G. Abell, M. Honda, A. Carroll, M. Gartland, B. Clarke, K. Suzuki, R. Lanford, D. V. Sangar, and S. M. Lemon. 1999. An infectious molecular clone of a Japanese genotype 1b hepatitis C virus. Hepatology 30:316-324. - PubMed
-
- Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. - PubMed
-
- De Francesco, R., P. Neddermann, L. Tomei, C. Steinkuhler, P. Gallinari, and A. Folgori. 2000. Biochemical and immunologic properties of the nonstructural proteins of the hepatitis C virus: implications for development of antiviral agents and vaccines. Semin. Liver Dis. 20:69-83. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

