Processing of nucleopeptides mimicking the topoisomerase I-DNA covalent complex by tyrosyl-DNA phosphodiesterase
- PMID: 11861912
- PMCID: PMC101246
- DOI: 10.1093/nar/30.5.1198
Processing of nucleopeptides mimicking the topoisomerase I-DNA covalent complex by tyrosyl-DNA phosphodiesterase
Abstract
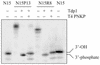
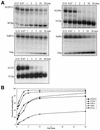
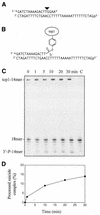
Tyrosyl-DNA phosphodiesterase-1 (Tdp1) is the only known enzyme to remove tyrosine from complexes in which the amino acid is linked to the 3'-end of DNA fragments. Such complexes can be produced following DNA processing by topoisomerase I, and recent studies in yeast have demonstrated the importance of TDP1 for cell survival following topoisomerase I-mediated DNA damage. In the present study, we used synthetic oligodeoxynucleotide-peptide conjugates (nucleopeptides) and recombinant yeast Tdp1 to investigate the molecular determinants for Tdp1 activity. We find that Tdp1 can process nucleopeptides with up to 13 amino acid residues but is poorly active with a 70 kDa fragment of topoisomerase I covalently linked to a suicide DNA substrate. Furthermore, Tdp1 was more effective with nucleopeptides with one to four amino acids than 15 amino acids. Tdp1 was also more effective with nucleopeptides containing 15 nt than with homolog nucleopeptides containing 4 nt. These results suggest that DNA binding contributes to the activity of Tdp1 and that Tdp1 would be most effective after topoisomerase I has been proteolyzed in vivo.
Figures


Similar articles
-
Pathways for repair of topoisomerase I covalent complexes in Saccharomyces cerevisiae.Genes Cells. 2001 Aug;6(8):677-87. doi: 10.1046/j.1365-2443.2001.00452.x. Genes Cells. 2001. PMID: 11532027
-
Tyrosyl-DNA phosphodiesterase (Tdp1) participates in the repair of Top2-mediated DNA damage.Proc Natl Acad Sci U S A. 2006 Jun 13;103(24):8953-8. doi: 10.1073/pnas.0603455103. Epub 2006 Jun 2. Proc Natl Acad Sci U S A. 2006. PMID: 16751265 Free PMC article.
-
A eukaryotic enzyme that can disjoin dead-end covalent complexes between DNA and type I topoisomerases.Proc Natl Acad Sci U S A. 1996 Oct 15;93(21):11534-9. doi: 10.1073/pnas.93.21.11534. Proc Natl Acad Sci U S A. 1996. PMID: 8876170 Free PMC article.
-
Tyrosyl-DNA phosphodiesterase I resolves both naturally and chemically induced DNA adducts and its potential as a therapeutic target.Drug Metab Rev. 2014 Nov;46(4):494-507. doi: 10.3109/03602532.2014.971957. Epub 2014 Oct 20. Drug Metab Rev. 2014. PMID: 25327705 Review.
-
Natural Products and Their Derivatives as Inhibitors of the DNA Repair Enzyme Tyrosyl-DNA Phosphodiesterase 1.Int J Mol Sci. 2023 Mar 17;24(6):5781. doi: 10.3390/ijms24065781. Int J Mol Sci. 2023. PMID: 36982848 Free PMC article. Review.
Cited by
-
The DNA binding and 3'-end preferential activity of human tyrosyl-DNA phosphodiesterase.Nucleic Acids Res. 2010 Apr;38(7):2444-52. doi: 10.1093/nar/gkp1206. Epub 2010 Jan 21. Nucleic Acids Res. 2010. PMID: 20097655 Free PMC article.
-
Ubx5-Cdc48 assists the protease Wss1 at DNA-protein crosslink sites in yeast.EMBO J. 2023 Jul 3;42(13):e113609. doi: 10.15252/embj.2023113609. Epub 2023 May 5. EMBO J. 2023. PMID: 37144685 Free PMC article.
-
Structure Modeling of Human Tyrosyl-DNA Phosphodiesterase 1 and Screening for Its Inhibitors.Acta Naturae. 2017 Apr-Jun;9(2):59-66. Acta Naturae. 2017. PMID: 28740727 Free PMC article.
-
Effects of DNA and protein size on substrate cleavage by human tyrosyl-DNA phosphodiesterase 1.Biochem J. 2011 Jun 15;436(3):559-66. doi: 10.1042/BJ20101841. Biochem J. 2011. PMID: 21463258 Free PMC article.
-
Application of a bivalent "click" approach to target tyrosyl-DNA phosphodiesterase 1 (TDP1).RSC Med Chem. 2025 Feb 21;16(5):1969-1985. doi: 10.1039/d4md00824c. eCollection 2025 May 22. RSC Med Chem. 2025. PMID: 39990162 Free PMC article.
References
-
- Wang J.C. (1996) DNA topoisomerases. Annu. Rev. Biochem., 65, 635–692. - PubMed
-
- Champoux J.J. (2001) DNA topoisomerases: structure, function and mechanism. Annu. Rev. Biochem., 70, 369–413. - PubMed
-
- Wigley D.B. (1998) Teaching a new dog old tricks? Curr. Biol., 6, 543–548. - PubMed
-
- Cheng C., Kussie,P., Pavletich,N. and Shuman,S. (1998) Conservation of structure and mechanism between eukaryotic topoisomerase I and site-specific recombinases. Cell, 92, 841–850. - PubMed
-
- Pourquier P. and Pommier,Y. (2001) Topoisomerase I-mediated DNA damage. Adv. Cancer Res., 80, 189–216. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

