Identification and comparative analysis of the chloroplast alpha-subunit gene of DNA-dependent RNA polymerase from seven Euglena species
- PMID: 11861918
- PMCID: PMC101230
- DOI: 10.1093/nar/30.5.1247
Identification and comparative analysis of the chloroplast alpha-subunit gene of DNA-dependent RNA polymerase from seven Euglena species
Abstract
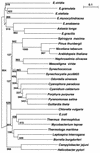
When the sequence of the Euglena gracilis chloroplast genome was reported in 1993 the alpha-subunit gene (rpoA) of RNA polymerase appeared to be missing, based on a comparison of all putative reading frames to the then known rpoA loci. Since there has been a large increase in known rpoA sequences, the question of a Euglena chloroplast rpoA gene was re-examined. A previously described unknown reading frame of 161 codons was found to be part of an rpoA gene split by a single group III intron. This rpoA gene, which is highly variable from species to species, was then isolated and characterized in five other euglenoid species, Euglena anabaena, Euglena granulata, Euglena myxocylindracea, Euglena stellata and Euglena viridis, and in the Astasia longa plastid genome. All seven Euglena rpoA genes have either one or three group III introns. The rpoA gene products in Euglena spp. appear to be the most variable in this gene family when compared to the rpoA gene in other species of bacteria, algae and plants. Additionally, Euglena rpoA proteins lack a C-terminal domain required for interaction with some regulatory proteins, a feature shared only with some chlorophyte green algae. The E.gracilis rpoA gene is the distal cistron of a multigene cluster that includes genes for carbohydrate biosynthesis, photosynthetic electron transport, an antenna complex and ribosomal proteins. This study provides new insights into the transcription system of euglenoid plastids, the organization of the plastid genome, group III intron evolution and euglenoid phylogeny.
Figures




References
-
- Greenberg B.M., Narita,J.O., DeLuca-Flaherty,C., Gruissem,W., Rushlow,K.A. and Hallick,R.B. (1984) Evidence for two RNA polymerase activities in Euglena gracilis chloroplasts. J. Biol. Chem., 259, 14880–14887. - PubMed
-
- Hess W.R. and Borner,T. (1999) Organellar RNA polymerases of higher plants. Int. Rev. Cytol., 190, 1–59. - PubMed
-
- Santis-MacIossek G., Kofer,W., Bock,A., Schoch,S., Maier,R.M., Wanner,G., Rudiger,W., Koop,H.U. and Herrmann,R.G. (1999) Targeted disruption of the plastid RNA polymerase genes rpoA, B and C1: molecular biology, biochemistry and ultrastructure. Plant J., 18, 477–489. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

