Exploring prokaryotic diversity in the genomic era
- PMID: 11864374
- PMCID: PMC139013
- DOI: 10.1186/gb-2002-3-2-reviews0003
Exploring prokaryotic diversity in the genomic era
Abstract
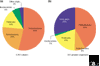
Our understanding of prokaryote biology from study of pure cultures and genome sequencing has been limited by a pronounced sampling bias towards four bacterial phyla - Proteobacteria, Firmicutes, Actinobacteria and Bacteroidetes - out of 35 bacterial and 18 archaeal phylum-level lineages. This bias is beginning to be rectified by the use of phylogenetically directed isolation strategies and by directly accessing microbial genomes from environmental samples.
Figures

References
-
- Staley JT, Konopka A. Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annu Rev Microbiol. 1985;39:321–346. - PubMed
-
- Galvez A, Maqueda M, Martinez-Bueno M, Valdivia E. Publication rates reveal trends in microbiological research. ASM News. 1998;64:269–275.
-
- DSMZ Bacterial Nomenclature Up-to-date http://www.dsmz.de/bactnom/bactname.htm
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

