14-3-3 transits to the nucleus and participates in dynamic nucleocytoplasmic transport
- PMID: 11864996
- PMCID: PMC2173313
- DOI: 10.1083/jcb.200112059
14-3-3 transits to the nucleus and participates in dynamic nucleocytoplasmic transport
Erratum in
- J Cell Biol 2002 Apr 29;157(3):533
Abstract
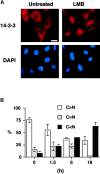
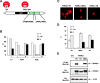
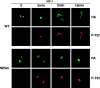
14-3-3 proteins regulate the cell cycle and prevent apoptosis by controlling the nuclear and cytoplasmic distribution of signaling molecules with which they interact. Although the majority of 14-3-3 molecules are present in the cytoplasm, we show here that in the absence of bound ligands 14-3-3 homes to the nucleus. We demonstrate that phosphorylation of one important 14-3-3 binding molecule, the transcription factor FKHRL1, at the 14-3-3 binding site occurs within the nucleus immediately before FKHRL1 relocalization to the cytoplasm. We show that the leucine-rich region within the COOH-terminal alpha-helix of 14-3-3, which had been proposed to function as a nuclear export signal (NES), instead functions globally in ligand binding and does not directly mediate nuclear transport. Efficient nuclear export of FKHRL1 requires both intrinsic NES sequences within FKHRL1 and phosphorylation/14-3-3 binding. Finally, we present evidence that phosphorylation/14-3-3 binding may also prevent FKHRL1 nuclear reimport. These results indicate that 14-3-3 can mediate the relocalization of nuclear ligands by several mechanisms that ensure complete sequestration of the bound 14-3-3 complex in the cytoplasm.
Figures





References
-
- Beck, T., and M.N. Hall. 1999. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature. 402:689–692. - PubMed
-
- Brunet, A., A. Bonni, M.J. Zigmond, M.Z. Lin, P. Juo, L.S. Hu, M.J. Anderson, K.C. Arden, J. Blenis, and M.E. Greenberg. 1999. Akt promotes cell survival by phosphorylating and inhibiting a forkhead transcription factor. Cell. 96:857–868. - PubMed
-
- Cahill, C.M., G. Tzivion, N. Nasrin, S. Ogg, J. Dore, G. Ruvkun, and M. Alexander-Bridges. 2001. Phosphatidylinositol 3-kinase signaling inhibits DAF-16 DNA binding and function via 14-3-3-dependent and 14-3-3-independent pathways. J. Biol. Chem. 276:13402–13410. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

