Control of intracellular dynamics of mammalian period proteins by casein kinase I epsilon (CKIepsilon) and CKIdelta in cultured cells
- PMID: 11865049
- PMCID: PMC135601
- DOI: 10.1128/MCB.22.6.1693-1703.2002
Control of intracellular dynamics of mammalian period proteins by casein kinase I epsilon (CKIepsilon) and CKIdelta in cultured cells
Abstract
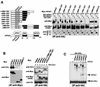
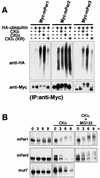
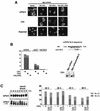
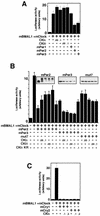
Recent studies have shown that casein kinase I epsilon (CKIepsilon) is an essential regulator of the mammalian circadian clock. However, the detailed mechanisms by which CKIepsilon regulates each component of the circadian negative-feedback loop have not been fully defined. We show here that mPer proteins, negative limbs of the autoregulatory loop, are specific substrates for CKIepsilon and CKIdelta. The CKI phosphorylation of mPer1 and mPer3 proteins results in their rapid degradation, which is dependent on the ubiquitin-proteasome pathway. Moreover, CKIepsilon and CKIdelta are able to induce nuclear translocation of mPer3, which requires its nuclear localization signal. The mutation in potential phosphorylation sites on mPer3 decreased the extent of both nuclear translocation and degradation of mPer3 that are stimulated by CKIepsilon. CKIepsilon and CKIdelta affected the inhibitory effect of mPer proteins on the transcriptional activity of BMAL1-CLOCK, but the inhibitory effect of mCry proteins on the activity of BMAL1-CLOCK was unaffected. These results suggest that CKIepsilon and CKIdelta regulate the mammalian circadian autoregulatory loop by controlling both protein turnover and subcellular localization of mPer proteins.
Figures



References
-
- Albrecht, U., Z. S. Sun, G. Eichele, and C. C. Lee. 1997. A differential response of two putative mammalian circadian regulators, mPer1 and mPer2, to light. Cell 91:1055-1064. - PubMed
-
- Brown, S. A., and U. Schibler. 1999. The ins and outs of circadian timekeeping. Curr. Opin. Genet. Dev. 9:588-594. - PubMed
-
- Darlington, T. K., K. Wager-Smith, M. F. Ceriani, D. Staknis, N. Gekakis, T. D. Steeves, C. J. Weitz, J. S. Takahashi, and S. A. Kay. 1998. Closing the circadian loop: CLOCK-induced transcription of its own inhibitors per and tim. Science 280:1599-1603. - PubMed
-
- Devlin, P. F., and S. A. Kay. 1999. Cryptochromes—bringing the blues to circadian rhythms. Trends Cell Biol. 9:295-298. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
