The rhesus rotavirus VP4 sialic acid binding domain has a galectin fold with a novel carbohydrate binding site
- PMID: 11867517
- PMCID: PMC125907
- DOI: 10.1093/emboj/21.5.885
The rhesus rotavirus VP4 sialic acid binding domain has a galectin fold with a novel carbohydrate binding site
Abstract
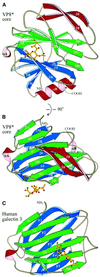
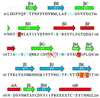
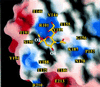
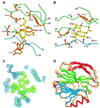
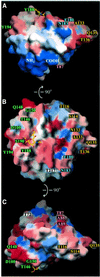
Cell attachment and membrane penetration are functions of the rotavirus outer capsid spike protein, VP4. An activating tryptic cleavage of VP4 produces the N-terminal fragment, VP8*, which is the viral hemagglutinin and an important target of neutralizing antibodies. We have determined, by X-ray crystallography, the atomic structure of the VP8* core bound to sialic acid and, by NMR spectroscopy, the structure of the unliganded VP8* core. The domain has the beta-sandwich fold of the galectins, a family of sugar binding proteins. The surface corresponding to the galectin carbohydrate binding site is blocked, and rotavirus VP8* instead binds sialic acid in a shallow groove between its two beta-sheets. There appears to be a small induced fit on binding. The residues that contact sialic acid are conserved in sialic acid-dependent rotavirus strains. Neutralization escape mutations are widely distributed over the VP8* surface and cluster in four epitopes. From the fit of the VP8* core into the virion spikes, we propose that VP4 arose from the insertion of a host carbohydrate binding domain into a viral membrane interaction protein.
Figures


References
-
- Bartels C., Xia,T.-H., Billeter,M., Guntert,P. and Wuthrich,K. (1995) The program XEASY for computer-supported NMR spectral analysis of biological macromolecules. J. Biomol. NMR, 5, 1–10. - PubMed
-
- Brunger A.T. et al. (1998) Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. - PubMed
-
- CCP4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. - PubMed
-
- Centers for Disease Control (1999) Withdrawal of rotavirus vaccine recommendation. MMWR Morb. Mortal. Wkly Rep., 48, 1007. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

