The SecYEG preprotein translocation channel is a conformationally dynamic and dimeric structure
- PMID: 11867527
- PMCID: PMC125904
- DOI: 10.1093/emboj/21.5.995
The SecYEG preprotein translocation channel is a conformationally dynamic and dimeric structure
Abstract
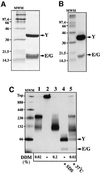
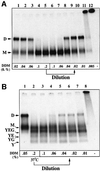
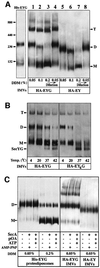
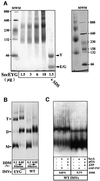
Escherichia coli preprotein translocase comprises a membrane-embedded trimeric complex of SecY, SecE and SecG. Previous studies have shown that this complex forms ring-like assemblies, which are thought to represent the preprotein translocation channel across the membrane. We have analyzed the functional state and the quaternary structure of the SecYEG translocase by employing cross-linking and blue native gel electrophoresis. The results show that the SecYEG monomer is a highly dynamic structure, spontaneously and reversibly associating into dimers. SecG-dependent tetramers and higher order SecYEG multimers can also exist in the membrane, but these structures form at high SecYEG concentration or upon overproduction of the complex only. The translocation process does not affect the oligomeric state of the translocase and arrested preproteins can be trapped with SecYEG or SecYE dimers. Dissociation of the dimer into a monomer by detergent induces release of the trapped preprotein. These results provide direct evidence that preproteins cross the bacterial membrane, associated with a translocation channel formed by a dimer of SecYEG.
Figures



References
-
- Beckmann R., Bubeck,D., Grassucci,R., Penczek,P., Verschoor,A., Blobel,G. and Frank,J. (1997) Alignment of conduits for the nascent polypeptide chain in the ribosome–Sec61 complex. Science, 278, 2123–2126. - PubMed
-
- Brundage L., Fimmel,C.J., Mizushima,S. and Wickner,W. (1992) SecY, SecE and band 1 form the membrane-embedded domain of Escherichia coli preprotein translocase. J. Biol. Chem., 267, 4166–4170. - PubMed
-
- Danese P.N. and Silhavy,T.J. (1998) Targeting and assembly of periplasmic and outer-membrane proteins in Escherichia coli. Annu. Rev. Genet., 32, 59–94. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

