The C. elegans ric-3 gene is required for maturation of nicotinic acetylcholine receptors
- PMID: 11867529
- PMCID: PMC125878
- DOI: 10.1093/emboj/21.5.1012
The C. elegans ric-3 gene is required for maturation of nicotinic acetylcholine receptors
Abstract
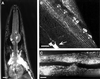
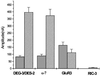
Mutations in ric-3 (resistant to inhibitors of cholinesterase) suppress the neuronal degenerations caused by a gain of function mutation in the Caenorhabditis elegans DEG-3 acetylcholine receptor. RIC-3 is a novel protein with two transmembrane domains and extensive coiled-coil domains. It is expressed in both muscles and neurons, and the protein is concentrated within the cell bodies. We demonstrate that RIC-3 is required for the function of at least four nicotinic acetylcholine receptors. However, GABA and glutamate receptors expressed in the same cells are unaffected. In ric-3 mutants, the DEG-3 receptor accumulates in the cell body instead of in the cell processes. Moreover, co-expression of ric-3 in Xenopus laevis oocytes enhances the activity of the C.elegans DEG-3/DES-2 and of the rat alpha-7 acetylcholine receptors. Together, these data suggest that RIC-3 is specifically required for the maturation of acetylcholine receptors.
Figures


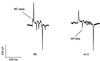


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

