Hyperacetylation of chromatin at the ADH2 promoter allows Adr1 to bind in repressed conditions
- PMID: 11867538
- PMCID: PMC125900
- DOI: 10.1093/emboj/21.5.1101
Hyperacetylation of chromatin at the ADH2 promoter allows Adr1 to bind in repressed conditions
Abstract
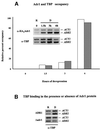
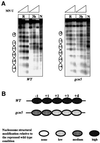
We report that in vivo increased acetylation of the repressed Saccharomyces cerevisiae ADH2 promoter chromatin, as obtained by disrupting the genes for the two deacetylases HDA1 and RPD3, destabilizes the structure of the TATA box-containing nucleosome. This acetylation-dependent chromatin remodeling is not sufficient to allow the binding of the TATA box-binding protein, but facilitates the recruitment of the transcriptional activator Adr1 and induces faster kinetics of mRNA accumulation when the cells are shifted to derepressing conditions.
Figures






Similar articles
-
Post-TATA binding protein recruitment clearance of Gcn5-dependent histone acetylation within promoter nucleosomes.Mol Cell Biol. 2003 Nov;23(21):7809-17. doi: 10.1128/MCB.23.21.7809-7817.2003. Mol Cell Biol. 2003. PMID: 14560024 Free PMC article.
-
A transient histone hyperacetylation signal marks nucleosomes for remodeling at the PHO8 promoter in vivo.Mol Cell. 2001 Mar;7(3):529-38. doi: 10.1016/s1097-2765(01)00200-3. Mol Cell. 2001. PMID: 11463378
-
Common chromatin architecture, common chromatin remodeling, and common transcription kinetics of Adr1-dependent genes in Saccharomyces cerevisiae.Biochemistry. 2004 Jul 13;43(27):8878-84. doi: 10.1021/bi049577+. Biochemistry. 2004. PMID: 15236596
-
A SAGA of histone acetylation and gene expression.Trends Genet. 1997 Nov;13(11):427-9. doi: 10.1016/s0168-9525(97)01292-4. Trends Genet. 1997. PMID: 9385836 Review. No abstract available.
-
Recruitment of chromatin remodelling factors during gene activation via the glucocorticoid receptor N-terminal domain.Biochem Soc Trans. 2000;28(4):410-4. Biochem Soc Trans. 2000. PMID: 10961930 Review.
Cited by
-
Combined global localization analysis and transcriptome data identify genes that are directly coregulated by Adr1 and Cat8.Mol Cell Biol. 2005 Mar;25(6):2138-46. doi: 10.1128/MCB.25.6.2138-2146.2005. Mol Cell Biol. 2005. PMID: 15743812 Free PMC article.
-
Repressive chromatin affects factor binding at yeast HO (homothallic switching) promoter.J Biol Chem. 2011 Oct 7;286(40):34809-19. doi: 10.1074/jbc.M111.281626. Epub 2011 Aug 12. J Biol Chem. 2011. PMID: 21840992 Free PMC article.
-
The Saccharomyces cerevisiae acetyltransferase Gcn5 exerts antagonistic pleiotropic effects on chronological ageing.Aging (Albany NY). 2023 Oct 23;15(20):10915-10937. doi: 10.18632/aging.205109. Epub 2023 Oct 23. Aging (Albany NY). 2023. PMID: 37874684 Free PMC article.
-
The transcriptional coactivators SAGA, SWI/SNF, and mediator make distinct contributions to activation of glucose-repressed genes.J Biol Chem. 2008 Nov 28;283(48):33101-9. doi: 10.1074/jbc.M805258200. Epub 2008 Sep 30. J Biol Chem. 2008. PMID: 18826948 Free PMC article.
-
Natural promoters and promoter engineering strategies for metabolic regulation in Saccharomyces cerevisiae.J Ind Microbiol Biotechnol. 2023 Feb 17;50(1):kuac029. doi: 10.1093/jimb/kuac029. J Ind Microbiol Biotechnol. 2023. PMID: 36633543 Free PMC article. Review.
References
-
- Aalfs J.D. and Kingston,R.E. (2000) What does ‘chromatin remodeling’ mean? Trends Biochem. Sci., 25, 548–555. - PubMed
-
- Anderson J.D., Lowary,P.T. and Widom,J. (2001) Effects of histone acetylation on the equilibrium accessibility of nucleosomal DNA target sites. J. Mol. Biol., 307, 977–985. - PubMed
-
- Beier D.R. and Young,E.T. (1982) Characterization of a regulatory region upstream of the ADR2 locus of S. cerevisiae. Nature, 300, 724–728. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

