Resolution of Prochlorococcus and Synechococcus ecotypes by using 16S-23S ribosomal DNA internal transcribed spacer sequences
- PMID: 11872466
- PMCID: PMC123739
- DOI: 10.1128/AEM.68.3.1180-1191.2002
Resolution of Prochlorococcus and Synechococcus ecotypes by using 16S-23S ribosomal DNA internal transcribed spacer sequences
Abstract
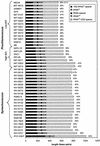
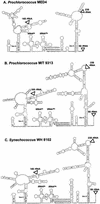
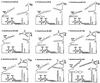
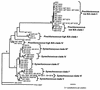
Cultured isolates of the marine cyanobacteria Prochlorococcus and Synechococcus vary widely in their pigment compositions and growth responses to light and nutrients, yet show greater than 96% identity in their 16S ribosomal DNA (rDNA) sequences. In order to better define the genetic variation that accompanies their physiological diversity, sequences for the 16S-23S rDNA internal transcribed spacer (ITS) region were determined in 32 Prochlorococcus isolates and 25 Synechococcus isolates from around the globe. Each strain examined yielded one ITS sequence that contained two tRNA genes. Dramatic variations in the length and G+C content of the spacer were observed among the strains, particularly among Prochlorococcus strains. Secondary-structure models of the ITS were predicted in order to facilitate alignment of the sequences for phylogenetic analyses. The previously observed division of Prochlorococcus into two ecotypes (called high and low-B/A after their differences in chlorophyll content) were supported, as was the subdivision of the high-B/A ecotype into four genetically distinct clades. ITS-based phylogenies partitioned marine cluster A Synechococcus into six clades, three of which can be associated with a particular phenotype (motility, chromatic adaptation, and lack of phycourobilin). The pattern of sequence divergence within and between clades is suggestive of a mode of evolution driven by adaptive sweeps and implies that each clade represents an ecologically distinct population. Furthermore, many of the clades consist of strains isolated from disparate regions of the world's oceans, implying that they are geographically widely distributed. These results provide further evidence that natural populations of Prochlorococcus and Synechococcus consist of multiple coexisting ecotypes, genetically closely related but physiologically distinct, which may vary in relative abundance with changing environmental conditions.
Figures

References
-
- Apirion, D., and A. Miczak. 1993. RNA processing in prokaryotic cells. BioEssays 15:113-120. - PubMed
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1992. Short protocols in molecular biology. John Wiley & Sons, New York, N.Y.
-
- Barry, T., G. Colleran, M. Glennon, L. R. Dunican, and F. Gannon. 1991. The 16S/23S ribosomal spacer region as a target for DNA probes to identify eubacteria. PCR Methods Appl. 1:51-56. - PubMed
-
- Béjà, O., M. T. Suzuki, E. V. Koonin, L. Aravind, A. Hadd, L. P. Nguyen, R. Villacorta, M. Amjadi, C. Garrigues, S. B. Jovanovich, R. A. Feldman, and E. F. DeLong. 2000. Construction and analysis of bacterial artificial chromosome libraries from a marine microbial assemblage. Environ. Microbiol. 2:516-529. - PubMed
-
- Berg, K. L., C. Squires, and C. L. Squires. 1989. Ribosomal RNA operon antitermination. Function of leader and spacer region box B-box A sequences and their conservation in diverse microorganisms. J. Mol. Biol. 209:345-358. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

