Wax ester production from n-alkanes by Acinetobacter sp. strain M-1: ultrastructure of cellular inclusions and role of acyl coenzyme A reductase
- PMID: 11872467
- PMCID: PMC123770
- DOI: 10.1128/AEM.68.3.1192-1195.2002
Wax ester production from n-alkanes by Acinetobacter sp. strain M-1: ultrastructure of cellular inclusions and role of acyl coenzyme A reductase
Abstract
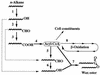
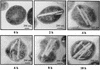
Acinetobacter sp. strain M-1 accumulated a large amount of wax esters from an n-alkane under nitrogen-limiting conditions. Under the optimized conditions with n-hexadecane as the substrate, the amount of hexadecyl hexadecanoate in the cells reached 0.17 g/g of cells (dry weight). Electron microscopic analysis revealed that multilayered disk-shaped intracellular inclusions were formed concomitant with wax ester formation. The contribution of acyl-CoA reductase to wax ester synthesis was evaluated by gene disruption analysis.
Figures


References
-
- Dewitt, S., J. L. Ervin, D. Howes-Orchison, D. Dalietos, and S. L. Neidleman. 1982. Saturated and unsaturated wax esters produced by Acinetobacter sp. HO1-N grown on C16-C20 n-alkanes. J. Am. Oil Chemists Soc. 59:69-74.
-
- Fixter, L. M., M. N. Nagi, J. G. McCormack, and C. A. Fewson. 1986. Structure, distribution and function of wax esters in Acinetobacter calcoaceticus. J. Gen. Microbiol. 132:3147-3157.
-
- Heuser, J. 1981. Preparing biological samples for stereo-microscopy by the quick-freeze, deep-etch, rotary-replication technique. Methods Cell Biol. 22:97-122. - PubMed
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

