Osmoregulation of dimer resolution at the plasmid pJHCMW1 mwr locus by Escherichia coli XerCD recombination
- PMID: 11872712
- PMCID: PMC134880
- DOI: 10.1128/JB.184.6.1607-1616.2002
Osmoregulation of dimer resolution at the plasmid pJHCMW1 mwr locus by Escherichia coli XerCD recombination
Abstract
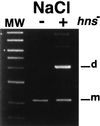
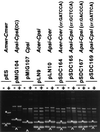
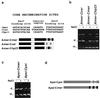
Xer-mediated dimer resolution at the mwr site of plasmid pJHCMW1 is osmoregulated in Escherichia coli. Whereas under low-salt conditions, the site-specific recombination reaction is efficient, under high-salt conditions, it proceeds inefficiently. Regulation of dimer resolution is independent of H-NS and is mediated by changes in osmolarity rather than ionic effects. The low level of recombination at high salt concentrations can be overcome by high levels of PepA or by mutating the ARG box to a sequence closer to the E. coli ARG box consensus. The central region of the mwr core recombination site plays a role in regulation of site-specific recombination by the osmotic pressure of the medium.
Figures

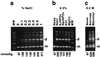




Similar articles
-
Stability by multimer resolution of pJHCMW1 is due to the Tn1331 resolvase and not to the Escherichia coli Xer system.Microbiology (Reading). 2000 Mar;146 ( Pt 3):581-589. doi: 10.1099/00221287-146-3-581. Microbiology (Reading). 2000. PMID: 10746761
-
Differences in resolution of mwr-containing plasmid dimers mediated by the Klebsiella pneumoniae and Escherichia coli XerC recombinases: potential implications in dissemination of antibiotic resistance genes.J Bacteriol. 2006 Apr;188(8):2812-20. doi: 10.1128/JB.188.8.2812-2820.2006. J Bacteriol. 2006. PMID: 16585742 Free PMC article.
-
mwr Xer site-specific recombination is hypersensitive to DNA supercoiling.Nucleic Acids Res. 2009 Jun;37(11):3580-7. doi: 10.1093/nar/gkp208. Epub 2009 Apr 9. Nucleic Acids Res. 2009. PMID: 19359357 Free PMC article.
-
Timing, self-control and a sense of direction are the secrets of multicopy plasmid stability.Mol Microbiol. 1998 Sep;29(5):1137-45. doi: 10.1046/j.1365-2958.1998.01012.x. Mol Microbiol. 1998. PMID: 9767582 Review.
-
The topology of plasmid-monomerizing Xer site-specific recombination.Biochem Soc Trans. 2013 Apr;41(2):589-94. doi: 10.1042/BST20120340. Biochem Soc Trans. 2013. PMID: 23514159 Review.
Cited by
-
Functional Analysis of the Acinetobacter baumannii XerC and XerD Site-Specific Recombinases: Potential Role in Dissemination of Resistance Genes.Antibiotics (Basel). 2020 Jul 13;9(7):405. doi: 10.3390/antibiotics9070405. Antibiotics (Basel). 2020. PMID: 32668667 Free PMC article.
-
Small Klebsiella pneumoniae Plasmids: Neglected Contributors to Antibiotic Resistance.Front Microbiol. 2019 Sep 20;10:2182. doi: 10.3389/fmicb.2019.02182. eCollection 2019. Front Microbiol. 2019. PMID: 31616398 Free PMC article.
-
fpr, a deficient Xer recombination site from a Salmonella plasmid, fails to confer stability by dimer resolution: comparative studies with the pJHCMW1 mwr site.J Bacteriol. 2010 Feb;192(3):883-7. doi: 10.1128/JB.01082-09. Epub 2009 Dec 4. J Bacteriol. 2010. PMID: 19966005 Free PMC article.
-
Plasmid-Mediated Antibiotic Resistance and Virulence in Gram-negatives: the Klebsiella pneumoniae Paradigm.Microbiol Spectr. 2014;2(5):1-15. doi: 10.1128/microbiolspec.PLAS-0016-2013. Microbiol Spectr. 2014. PMID: 25705573 Free PMC article.
-
Complete nucleotide sequence of Klebsiella pneumoniae multiresistance plasmid pJHCMW1.Antimicrob Agents Chemother. 2002 Nov;46(11):3422-7. doi: 10.1128/AAC.46.11.3422-3427.2002. Antimicrob Agents Chemother. 2002. PMID: 12384346 Free PMC article.
References
-
- Arciszewska, L. K., R. A. Baker, B. Hallet, and D. J. Sherratt. 2000. Coordinated control of XerC and XerD catalytic activities during Holliday junction resolution. J. Mol. Biol. 299:391-403. - PubMed
-
- Blake, J. A., N. Ganguly, and D. J. Sherratt. 1997. DNA sequence of recombinase-binding sites can determine Xer site-specific recombination outcome. Mol. Microbiol. 23:387-398. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

