Glyoxylate regeneration pathway in the methylotroph Methylobacterium extorquens AM1
- PMID: 11872727
- PMCID: PMC134890
- DOI: 10.1128/JB.184.6.1750-1758.2002
Glyoxylate regeneration pathway in the methylotroph Methylobacterium extorquens AM1
Abstract
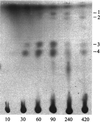
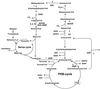
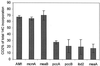
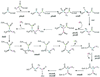
Most serine cycle methylotrophic bacteria lack isocitrate lyase and convert acetyl coenzyme A (acetyl-CoA) to glyoxylate via a novel pathway thought to involve butyryl-CoA and propionyl-CoA as intermediates. In this study we have used a genome analysis approach followed by mutation to test a number of genes for involvement in this novel pathway. We show that methylmalonyl-CoA mutase, an R-specific crotonase, isobutyryl-CoA dehydrogenase, and a GTPase are involved in glyoxylate regeneration. We also monitored the fate of (14)C-labeled carbon originating from acetate, butyrate, or bicarbonate in mutants defective in glyoxylate regeneration and identified new potential intermediates in the pathway: ethylmalonyl-CoA, methylsuccinyl-CoA, isobutyryl-CoA, methacrylyl-CoA, and beta-hydroxyisobutyryl-CoA. A new scheme for the pathway is proposed based on these data.
Figures

References
-
- Anthony, C. 1982. The biochemistry of methylotrophs. Academic Press, London, United Kingdom.
-
- Bellion, E., and J. C. Spain. 1975. The distribution of the isocitrate lyase serine pathway amongst one-carbon utilizing organisms. Can. J. Microbiol. 22:404-408. - PubMed
-
- Bobik, T. A., and M. E. Rasche. 2001. Identification of the human methylmalonyl-CoA racemase gene based on the analysis of prokaryotic gene arrangements: implications for decoding the human genome. J. Biol. Chem. 276:37194-37198. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

