Extraction of functional binding sites from unique regulatory regions: the Drosophila early developmental enhancers
- PMID: 11875036
- PMCID: PMC155290
- DOI: 10.1101/gr.212502
Extraction of functional binding sites from unique regulatory regions: the Drosophila early developmental enhancers
Abstract
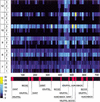
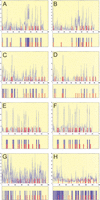
The early developmental enhancers of Drosophila melanogaster comprise one of the most sophisticated regulatory systems in higher eukaryotes. An elaborate code in their DNA sequence translates both maternal and early embryonic regulatory signals into spatial distribution of transcription factors. One of the most striking features of this code is the redundancy of binding sites for these transcription factors (BSTF). Using this redundancy, we explored the possibility of predicting functional binding sites in a single enhancer region without any prior consensus/matrix description or evolutionary sequence comparisons. We developed a conceptually simple algorithm, Scanseq, that employs an original statistical evaluation for identifying the most redundant motifs and locates the position of potential BSTF in a given regulatory region. To estimate the biological relevance of our predictions, we built thorough literature-based annotations for the best-known Drosophila developmental enhancers and we generated detailed distribution maps for the most robust binding sites. The high statistical correlation between the location of BSTF in these experiment-based maps and the location predicted in silico by Scanseq confirmed the relevance of our approach. We also discuss the definition of true binding sites and the possible biological principles that govern patterning of regulatory regions and the distribution of transcriptional signals.
Figures


References
-
- Andrioli LP, Vasisht V, Wasserman KT, Oberstein A, Kaplan L, Small S. 42nd Annual Drosophila Research Conference. 2001. The forkhead domain protein slp1 participates in combinatorial repression of even-skipped stripe 2. p. a37. The Genetics Society of America, Washington, D.C.
-
- Apostolico A, Bock ME, Lonardi S, Xu X. Efficient detection of unusual words. J Comput Biol. 2000;7:71–94. - PubMed
-
- Arnosti DN, Barolo S, Levine M, Small S. The eve stripe 2 enhancer employs multiple modes of transcriptional synergy. Development. 1996;122:205–214. - PubMed
-
- Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. - PubMed
-
- ————— Unsupervised learning of multiple motifs in biopolymers using expectation maximization. Machine Learning. 1995;21:51–80.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
