Hydrogenases and hydrogen metabolism of cyanobacteria
- PMID: 11875125
- PMCID: PMC120778
- DOI: 10.1128/MMBR.66.1.1-20.2002
Hydrogenases and hydrogen metabolism of cyanobacteria
Abstract
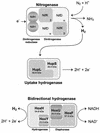
Cyanobacteria may possess several enzymes that are directly involved in dihydrogen metabolism: nitrogenase(s) catalyzing the production of hydrogen concomitantly with the reduction of dinitrogen to ammonia, an uptake hydrogenase (encoded by hupSL) catalyzing the consumption of hydrogen produced by the nitrogenase, and a bidirectional hydrogenase (encoded by hoxFUYH) which has the capacity to both take up and produce hydrogen. This review summarizes our knowledge about cyanobacterial hydrogenases, focusing on recent progress since the first molecular information was published in 1995. It presents the molecular knowledge about cyanobacterial hupSL and hoxFUYH, their corresponding gene products, and their accessory genes before finishing with an applied aspect--the use of cyanobacteria in a biological, renewable production of the future energy carrier molecular hydrogen. In addition to scientific publications, information from three cyanobacterial genomes, the unicellular Synechocystis strain PCC 6803 and the filamentous heterocystous Anabaena strain PCC 7120 and Nostoc punctiforme (PCC 73102/ATCC 29133) is included.
Figures

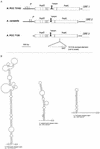


 represent the NAD- and flavin mononucleotide-binding regions, respectively.
represent the NAD- and flavin mononucleotide-binding regions, respectively.

 ). Anabaena strain PCC 7120 (A. PCC 7120) (, ;
). Anabaena strain PCC 7120 (A. PCC 7120) (, ; References
-
- Reference deleted.
-
- Reference deleted.
-
- Adams, D. G. 2000. Symbiotic interactions, p. 523-561. In B. A. Whitton and M. Potts (ed.), The ecology of cyanobacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
-
- Akhmanova, A., F. G. Voncken, K. M. Hosea, H. Harhangi, J. T. Keltjens, H. J. op den Camp, G. D. Vogels, and J. H. Hackstein. 1999. A hydrogenosome with pyruvate formate-lyase: anaerobic chytrid fungi use an alternative route for pyruvate catabolism. Mol. Microbiol. 32:1103-1114. - PubMed
-
- Akhmanova, A., F. G. Voncken, T. van Alen, A. van Hoek, B. Boxma, G. D. Vogels, M. Veenhuis, and J. H. Hackstein. 1998. A hydrogenosome with a genome. Nature 396:527-528. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

