Alpha-crystallin-type heat shock proteins: socializing minichaperones in the context of a multichaperone network
- PMID: 11875128
- PMCID: PMC120782
- DOI: 10.1128/MMBR.66.1.64-93.2002
Alpha-crystallin-type heat shock proteins: socializing minichaperones in the context of a multichaperone network
Abstract
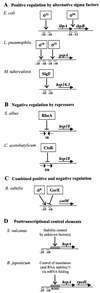
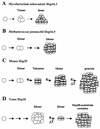
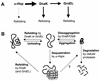
Alpha-crystallins were originally recognized as proteins contributing to the transparency of the mammalian eye lens. Subsequently, they have been found in many, but not all, members of the Archaea, Bacteria, and Eucarya. Most members of the diverse alpha-crystallin family have four common structural and functional features: (i) a small monomeric molecular mass between 12 and 43 kDa; (ii) the formation of large oligomeric complexes; (iii) the presence of a moderately conserved central region, the so-called alpha-crystallin domain; and (iv) molecular chaperone activity. Since alpha-crystallins are induced by a temperature upshift in many organisms, they are often referred to as small heat shock proteins (sHsps) or, more accurately, alpha-Hsps. Alpha-crystallins are integrated into a highly flexible and synergistic multichaperone network evolved to secure protein quality control in the cell. Their chaperone activity is limited to the binding of unfolding intermediates in order to protect them from irreversible aggregation. Productive release and refolding of captured proteins into the native state requires close cooperation with other cellular chaperones. In addition, alpha-Hsps seem to play an important role in membrane stabilization. The review compiles information on the abundance, sequence conservation, regulation, structure, and function of alpha-Hsps with an emphasis on the microbial members of this chaperone family.
Figures
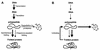



Similar articles
-
NMR spectroscopy of alpha-crystallin. Insights into the structure, interactions and chaperone action of small heat-shock proteins.Int J Biol Macromol. 1998 May-Jun;22(3-4):197-209. doi: 10.1016/s0141-8130(98)00017-8. Int J Biol Macromol. 1998. PMID: 9650074
-
Chaperone-like activity of alpha-crystallin and other small heat shock proteins.Curr Protein Pept Sci. 2001 Sep;2(3):205-25. doi: 10.2174/1389203013381107. Curr Protein Pept Sci. 2001. PMID: 12369933 Review.
-
Structure and function of small heat shock/alpha-crystallin proteins: established concepts and emerging ideas.Cell Mol Life Sci. 2000 Jun;57(6):899-913. doi: 10.1007/pl00000733. Cell Mol Life Sci. 2000. PMID: 10950306 Free PMC article. Review.
-
Molecular characterization of a small heat shock/alpha-crystallin protein in encysted Artemia embryos.J Biol Chem. 1997 Jul 25;272(30):19051-8. doi: 10.1074/jbc.272.30.19051. J Biol Chem. 1997. PMID: 9228089
-
Site-directed mutations within the core "alpha-crystallin" domain of the small heat-shock protein, human alphaB-crystallin, decrease molecular chaperone functions.J Mol Biol. 1999 Jun 4;289(2):397-411. doi: 10.1006/jmbi.1999.2759. J Mol Biol. 1999. PMID: 10366513
Cited by
-
Homeostatic Roles of the Proteostasis Network in Dendrites.Front Cell Neurosci. 2020 Aug 14;14:264. doi: 10.3389/fncel.2020.00264. eCollection 2020. Front Cell Neurosci. 2020. PMID: 33013325 Free PMC article. Review.
-
Identification and characterization of a stress-inducible and a constitutive small heat-shock protein targeted to the matrix of plant peroxisomes.Plant Physiol. 2006 May;141(1):47-60. doi: 10.1104/pp.105.073841. Epub 2006 Mar 10. Plant Physiol. 2006. PMID: 16531488 Free PMC article.
-
The function of the beta3 interactive domain in the small heat shock protein and molecular chaperone, human alphaB crystallin.Cell Stress Chaperones. 2006 Summer;11(2):187-97. doi: 10.1379/csc-186.1. Cell Stress Chaperones. 2006. PMID: 16817325 Free PMC article.
-
Mammalian Hsp22 is a heat-inducible small heat-shock protein with chaperone-like activity.Biochem J. 2004 Jul 15;381(Pt 2):379-87. doi: 10.1042/BJ20031958. Biochem J. 2004. PMID: 15030316 Free PMC article.
-
Substrate Protein Interactions and Methylglyoxal Modifications Reduce the Aggregation Propensity of Human Alpha-A-Crystallin G98R Mutant.Front Mol Biosci. 2022 Apr 6;9:875205. doi: 10.3389/fmolb.2022.875205. eCollection 2022. Front Mol Biosci. 2022. PMID: 35463950 Free PMC article.
References
-
- Abgar, S., N. Yevlampieva, T. Aerts, J. Vanhoudt, and J. Clauwaert. 2000. Chaperone-like activity of bovine lens α-crystallin in the presence of dithiothreitol-destabilized proteins: characterization of the formed complexes. Biochem. Biophys. Res. Commun. 276:619-625. - PubMed
-
- Abu Kwaik, Y., and N. C. Engleberg. 1994. Cloning and molecular characterization of a Legionella pneumophila gene induced by intracellular infection and by various in vitro stress conditions. Mol. Microbiol. 13:243-251. - PubMed
-
- Abu Kwaik, Y., L. Y. Gao, O. S. Harb, and B. J. Stone. 1997. Transcriptional regulation of the macrophage-induced gene (gspA) of Legionella pneumophila and phenotypic characterization of a null mutant. Mol. Microbiol. 24:629-642. - PubMed
-
- Akiyama, Y., M. Ehrmann, A. Kihara, and K. Ito. 1998. Polypeptide binding of Escherichia coli FtsH (HflB). Mol. Microbiol. 28:803-812. - PubMed
-
- Akiyama, Y., T. Ogura, and K. Ito. 1994. Involvement of FtsH in protein assembly into and through the membrane. I. Mutations that reduce retention of a cytoplasmic reporter. J. Biol. Chem. 269:5218-5224. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

