Correlating overrepresented upstream motifs to gene expression: a computational approach to regulatory element discovery in eukaryotes
- PMID: 11876822
- PMCID: PMC77394
- DOI: 10.1186/1471-2105-3-7
Correlating overrepresented upstream motifs to gene expression: a computational approach to regulatory element discovery in eukaryotes
Abstract
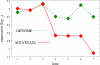
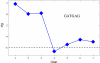
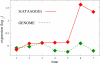
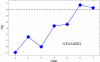
Background: Gene regulation in eukaryotes is mainly effected through transcription factors binding to rather short recognition motifs generally located upstream of the coding region. We present a novel computational method to identify regulatory elements in the upstream region of eukaryotic genes. The genes are grouped in sets sharing an overrepresented short motif in their upstream sequence. For each set, the average expression level from a microarray experiment is determined: If this level is significantly higher or lower than the average taken over the whole genome, then the overerpresented motif shared by the genes in the set is likely to play a role in their regulation.
Results: The method was tested by applying it to the genome of Saccharomyces cerevisiae, using the publicly available results of a DNA microarray experiment, in which expression levels for virtually all the genes were measured during the diauxic shift from fermentation to respiration. Several known motifs were correctly identified, and a new candidate regulatory sequence was determined.
Conclusions: We have described and successfully tested a simple computational method to identify upstream motifs relevant to gene regulation in eukaryotes by studying the statistical correlation between overepresented upstream motifs and gene expression levels.
Figures

References
-
- DeRisi JL, Iyer VR, Brown PO. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. http://cmgm.stanford.edu/pbrown/explore/ - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

