Arabidopsis COP10 is a ubiquitin-conjugating enzyme variant that acts together with COP1 and the COP9 signalosome in repressing photomorphogenesis
- PMID: 11877375
- PMCID: PMC155353
- DOI: 10.1101/gad.964602
Arabidopsis COP10 is a ubiquitin-conjugating enzyme variant that acts together with COP1 and the COP9 signalosome in repressing photomorphogenesis
Abstract
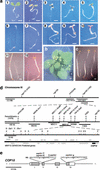
A group of evolutionarily conserved pleiotropic COP/DET/FUS proteins was initially defined by their ability to repress photomorphogenesis in Arabidopsis. It was proposed that this regulation be mediated by targeting degradation of key cellular regulators that promote photomorphogenesis. Among them, COP1 and the COP9 signalosome have been hypothesized to fulfill the roles as an ubiquitin ligase (E3) and an essential E3 modulator. Here we report that COP10 encodes a protein similar to ubiquitin-conjugating enzyme (E2) variant proteins (UEV). COP10 is part of a nuclear protein complex and capable of directly interacting with both COP1 and the COP9 signalosome. Our data indicates that COP10 defines a possible E2 activity, thus validating the working hypothesis that the pleiotropic COP/DET/FUS group of proteins defined a protein ubiquitination pathway.
Figures
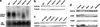



Similar articles
-
Arabidopsis CULLIN4 Forms an E3 Ubiquitin Ligase with RBX1 and the CDD Complex in Mediating Light Control of Development.Plant Cell. 2006 Aug;18(8):1991-2004. doi: 10.1105/tpc.106.043224. Epub 2006 Jul 14. Plant Cell. 2006. PMID: 16844902 Free PMC article.
-
Multiple ubiquitin ligase-mediated processes require COP9 signalosome and AXR1 function.Plant Cell. 2002 Oct;14(10):2553-63. doi: 10.1105/tpc.003434. Plant Cell. 2002. PMID: 12368504 Free PMC article.
-
The COP/DET/FUS proteins-regulators of eukaryotic growth and development.Semin Cell Dev Biol. 2000 Dec;11(6):495-503. doi: 10.1006/scdb.2000.0203. Semin Cell Dev Biol. 2000. PMID: 11145879 Review.
-
Arabidopsis COP10 forms a complex with DDB1 and DET1 in vivo and enhances the activity of ubiquitin conjugating enzymes.Genes Dev. 2004 Sep 1;18(17):2172-81. doi: 10.1101/gad.1229504. Genes Dev. 2004. PMID: 15342494 Free PMC article.
-
Constitutive photomorphogenesis protein 1 (COP1) and COP9 signalosome, evolutionarily conserved photomorphogenic proteins as possible targets of melatonin.J Pineal Res. 2016 Aug;61(1):41-51. doi: 10.1111/jpi.12340. Epub 2016 Jun 13. J Pineal Res. 2016. PMID: 27121162 Review.
Cited by
-
COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis.Plant Cell. 2008 Feb;20(2):292-306. doi: 10.1105/tpc.107.057281. Epub 2008 Feb 22. Plant Cell. 2008. PMID: 18296627 Free PMC article.
-
The Arabidopsis C2H2 zinc finger INDETERMINATE DOMAIN1/ENHYDROUS promotes the transition to germination by regulating light and hormonal signaling during seed maturation.Plant Cell. 2011 May;23(5):1772-94. doi: 10.1105/tpc.111.085134. Epub 2011 May 13. Plant Cell. 2011. PMID: 21571950 Free PMC article.
-
COP1 and phyB Physically Interact with PIL1 to Regulate Its Stability and Photomorphogenic Development in Arabidopsis.Plant Cell. 2014 Jun;26(6):2441-2456. doi: 10.1105/tpc.113.121657. Epub 2014 Jun 20. Plant Cell. 2014. PMID: 24951480 Free PMC article.
-
Roles of constitutive photomorphogenic 10 in Arabidopsis stomata development.Plant Signal Behav. 2012 Aug;7(8):990-3. doi: 10.4161/psb.20995. Epub 2012 Jul 27. Plant Signal Behav. 2012. PMID: 22836493 Free PMC article.
-
Arabidopsis CULLIN4 Forms an E3 Ubiquitin Ligase with RBX1 and the CDD Complex in Mediating Light Control of Development.Plant Cell. 2006 Aug;18(8):1991-2004. doi: 10.1105/tpc.106.043224. Epub 2006 Jul 14. Plant Cell. 2006. PMID: 16844902 Free PMC article.
References
-
- Ang LH, Chattopadhyay S, Wei N, Oyama T, Okada K, Batschauer A, Deng XW. Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsisdevelopment. Mol Cell. 1998;1:213–222. - PubMed
-
- Bell CJ, Ecker JR. Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics. 1994;19:137–144. - PubMed
-
- Deng L, Wang C, Spencer E, Yang L, Braun A, You J, Slaughter C, Pickart C, Chen ZJ. Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell. 2000;103:351–361. - PubMed
-
- Deshaies RJ. SCF and Cullin/Ring H2-based ubiquitin ligases. Annu Rev Cell Dev Biol. 1999;15:435–467. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
