The role of cell cycle-regulated expression in the localization of spatial landmark proteins in yeast
- PMID: 11877459
- PMCID: PMC2173311
- DOI: 10.1083/jcb.200107041
The role of cell cycle-regulated expression in the localization of spatial landmark proteins in yeast
Abstract
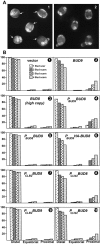
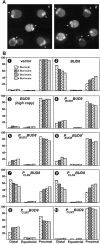
In Saccharomyces cerevisiae, Bud8p and Bud9p are homologous plasma membrane glycoproteins that appear to mark the distal and proximal cell poles, respectively, as potential sites for budding in the bipolar pattern. Here we provide evidence that Bud8p is delivered to the presumptive bud site (and thence to the distal pole of the bud) just before bud emergence, and that Bud9p is delivered to the bud side of the mother-bud neck (and thence to the proximal pole of the daughter cell) after activation of the mitotic exit network, just before cytokinesis. Like the delivery of Bud8p, that of Bud9p is actin dependent; unlike the delivery of Bud8p, that of Bud9p is also septin dependent. Interestingly, although the transcription of BUD8 and BUD9 appears to be cell cycle regulated, the abundance of BUD8 mRNA peaks in G2/M and that of BUD9 mRNA peaks in late G1, suggesting that the translation and/or delivery to the cell surface of each protein is delayed and presumably also cell cycle regulated. The importance of time of transcription in localization is supported by promoter-swap experiments: expression of Bud8p from the BUD9 promoter leads to its localization predominantly to the sites typical for Bud9p, and vice versa. Moreover, expression of Bud8p from the BUD9 promoter fails to rescue the budding-pattern defect of a bud8 mutant but fully rescues that of a bud9 mutant. However, although expression of Bud9p from the BUD8 promoter fails to rescue a bud9 mutant, it also rescues only partially the budding-pattern defect of a bud8 mutant, suggesting that some feature(s) of the Bud8p protein is also important for Bud8p function. Experiments with chimeric proteins suggest that the critical element(s) is somewhere in the extracytoplasmic domain of Bud8p.
Figures



Similar articles
-
Bud8p and Bud9p, proteins that may mark the sites for bipolar budding in yeast.Mol Biol Cell. 2001 Aug;12(8):2497-518. doi: 10.1091/mbc.12.8.2497. Mol Biol Cell. 2001. PMID: 11514631 Free PMC article.
-
Interactions among Rax1p, Rax2p, Bud8p, and Bud9p in marking cortical sites for bipolar bud-site selection in yeast.Mol Biol Cell. 2004 Nov;15(11):5145-57. doi: 10.1091/mbc.e04-07-0600. Epub 2004 Sep 8. Mol Biol Cell. 2004. PMID: 15356260 Free PMC article.
-
Asymmetrically localized Bud8p and Bud9p proteins control yeast cell polarity and development.EMBO J. 2000 Dec 15;19(24):6686-96. doi: 10.1093/emboj/19.24.6686. EMBO J. 2000. PMID: 11118203 Free PMC article.
-
Subcellular localization of the interaction of bipolar landmarks Bud8p and Bud9p with Rax2p in Saccharomyces cerevisiae diploid cells.Biochem Biophys Res Commun. 2010 Sep 3;399(4):525-30. doi: 10.1016/j.bbrc.2010.07.102. Epub 2010 Aug 1. Biochem Biophys Res Commun. 2010. PMID: 20678480
-
Distinct domains of yeast cortical tag proteins Bud8p and Bud9p confer polar localization and functionality.Mol Biol Cell. 2007 Sep;18(9):3323-39. doi: 10.1091/mbc.e06-10-0899. Epub 2007 Jun 20. Mol Biol Cell. 2007. PMID: 17581861 Free PMC article.
Cited by
-
Spatial landmarks regulate a Cdc42-dependent MAPK pathway to control differentiation and the response to positional compromise.Proc Natl Acad Sci U S A. 2016 Apr 5;113(14):E2019-28. doi: 10.1073/pnas.1522679113. Epub 2016 Mar 21. Proc Natl Acad Sci U S A. 2016. PMID: 27001830 Free PMC article.
-
The Path towards Predicting Evolution as Illustrated in Yeast Cell Polarity.Cells. 2020 Nov 24;9(12):2534. doi: 10.3390/cells9122534. Cells. 2020. PMID: 33255231 Free PMC article. Review.
-
Central roles of small GTPases in the development of cell polarity in yeast and beyond.Microbiol Mol Biol Rev. 2007 Mar;71(1):48-96. doi: 10.1128/MMBR.00028-06. Microbiol Mol Biol Rev. 2007. PMID: 17347519 Free PMC article. Review.
-
Spatial regulation of exocytosis and cell polarity: yeast as a model for animal cells.FEBS Lett. 2007 May 22;581(11):2119-24. doi: 10.1016/j.febslet.2007.03.043. Epub 2007 Mar 30. FEBS Lett. 2007. PMID: 17418146 Free PMC article. Review.
-
Asymmetric localization of flotillins/reggies in preassembled platforms confers inherent polarity to hematopoietic cells.Proc Natl Acad Sci U S A. 2003 Jul 8;100(14):8241-6. doi: 10.1073/pnas.1331629100. Epub 2003 Jun 25. Proc Natl Acad Sci U S A. 2003. PMID: 12826615 Free PMC article.
References
-
- Ausubel, F.M., R. Brent, R.E. Kingston, D.D. Moore, J.G. Seidman, J.A. Smith, and K. Struhl. 1995. Current Protocols in Molecular Biology. John Wiley & Sons, New York.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

