Specificity of tissue transglutaminase explains cereal toxicity in celiac disease
- PMID: 11877487
- PMCID: PMC2193762
- DOI: 10.1084/jem.20012028
Specificity of tissue transglutaminase explains cereal toxicity in celiac disease
Abstract
Celiac disease is caused by a selective lack of T cell tolerance for gluten. It is known that the enzyme tissue transglutaminase (tTG) is involved in the generation of T cell stimulatory gluten peptides through deamidation of glutamine, the most abundant amino acid in gluten. Only particular glutamine residues, however, are modified by tTG. Here we provide evidence that the spacing between glutamine and proline, the second most abundant amino acid in gluten, plays an essential role in the specificity of deamidation. On the basis of this, algorithms were designed and used to successfully predict novel T cell stimulatory peptides in gluten. Strikingly, these algorithms identified many similar peptides in the gluten-like hordeins from barley and secalins from rye but not in the avenins from oats. The avenins contain significantly lower percentages of proline residues, which offers a likely explanation for the lack of toxicity of oats. Thus, the unique amino acid composition of gluten and related proteins in barley and rye favors the generation of toxic T cell stimulatory gluten peptides by tTG. This provides a rationale for the observation that celiac disease patients are intolerant to these cereal proteins but not to other common food proteins.
Figures
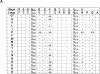

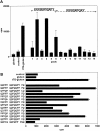
References
-
- Marsh, M.N. 1992. Gluten, major histocompatibility complex, and the small intestine. A molecular and immunobiologic approach to the spectrum of gluten sensitivity (‘celiac sprue’). Gastroenterology. 102:330–354. - PubMed
-
- Dieterich, W., T. Ehnis, M. Bauer, P. Donner, U. Volta, E.O. Riecken, and D. Schuppan. 1997. Identification of tissue transglutaminase as the autoantigen of celiac disease. Nat. Med. 3:797–801. - PubMed
-
- Molberg, O., S.N. McAdam, R. Korner, H. Quarsten, C. Kristiansen, L. Madsen, L. Fugger, H. Scott, O. Noren, P. Roepstorff, K.E. Lundin, H. Sjostrom, and L.M. Sollid. 1998. Tissue transglutaminase selectively modifies gliadin peptides that are recognized by gut-derived T cells in celiac disease. Nat. Med. 4:713–717. - PubMed
-
- van de Wal, Y., Y. Kooy, P. van Veelen, S. Pena, L. Mearin, G. Papadopoulos, and F. Koning. 1998. Selective deamidation by tissue transglutaminase strongly enhances gliadin-specific T cell reactivity. J. Immunol. 161:1585–1588. - PubMed
-
- Spurkland, A., G. Ingvarsson, E.S. Falk, I. Knutsen, L.M. Sollid, and E. Thorsby. 1997. Dermatitis herpetiformis and celiac disease are both primarily associated with the HLA-DQ (α1*0501, β1*02) or the HLA-DQ (α1*03, β1*0302) heterodimers. Tiss. Anttigens. 49:29–34. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

