Ca3 fingerprinting of Candida albicans isolates from human immunodeficiency virus-positive and healthy individuals reveals a new clade in South Africa
- PMID: 11880401
- PMCID: PMC120250
- DOI: 10.1128/JCM.40.3.826-836.2002
Ca3 fingerprinting of Candida albicans isolates from human immunodeficiency virus-positive and healthy individuals reveals a new clade in South Africa
Abstract
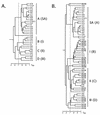
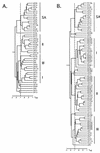
To examine the question of strain specificity in oropharyngeal candidiasis associated with human immunodeficiency virus (HIV) infection, oral samples were collected from 1,196 HIV-positive black South Africans visiting three clinics and 249 Candida albicans isolates were selected for DNA fingerprinting with the complex DNA fingerprinting probe Ca3. A total of 66 C. albicans isolates from healthy black South Africans and 46 from healthy white South Africans were also DNA fingerprinted as controls. Using DENDRON software, a cluster analysis was performed and the identified groups were compared to a test set of isolates from the United States in which three genetic groups (I, II, and III) were previously identified by a variety of genetic fingerprinting methods. All of the characterized South African collections (three from HIV-positive black persons, two from healthy black persons, and one from healthy white persons) included group I, II, and III isolates. In addition, all South African collections included a fourth group (group SA) completely absent in the U.S. collection. The proportion of group SA isolates in HIV-positive and healthy black South Africans was 53% in both cases. The proportion in healthy white South Africans was 33%. In a comparison of HIV-positive patients with and without oropharyngeal symptoms of infection, the same proportions of group I, II, III, and SA isolates were obtained, indicating no shift to a particular group on infection. However, by virtue of its predominance as a commensal and in infections, group SA must be considered the most successful in South Africa. Why group SA isolates represent 53 and 33% of colonizing strains in black and white South Africans and are absent in the U.S. collection represents an interesting epidemiological question.
Figures





References
-
- Arendorf, T. M., and D. M. Walker. 1980. The prevalence and intra-oral distribution of Candida albicans in man. Arch. Oral Biol. 25:1-10. - PubMed
-
- Brawner, D. L., and A. J. Hovan. 1995. Oral candidiasis in HIV-infected patients. Curr. Top. Med. Mycol. 6:113-125. - PubMed
-
- Coleman, D., D. Sullivan, B. Harrington, K. Haynes, M. Henman, D. Shanley, D. Bennett, G. Moran, C. McCreary, and L. O'Neill. 1997. Molecular and phenotypic analysis of Candida dubliniensis: a recently identified species linked with oral candidosis in HIV-infected and AIDS patients. Oral Dis. 3:S96-S101. - PubMed
-
- Coleman, D. C., D. E. Bennett, D. J. Sullivan, P. J. Gallagher, M. C. Henman, D. B. Shanley, and R. J. Russell. 1993. Oral Candida in HIV infection and AIDS: new perspectives/new approaches. Crit. Rev. Microbiol. 19:61-82. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

