Emulating biology: building nanostructures from the bottom up
- PMID: 11880609
- PMCID: PMC128548
- DOI: 10.1073/pnas.221458298
Emulating biology: building nanostructures from the bottom up
Abstract
The biological approach to nanotechnology has produced self-assembled objects, arrays and devices; likewise, it has achieved the recognition of inorganic systems and the control of their growth. Can these approaches now be integrated to produce useful systems?
Figures

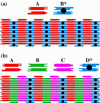
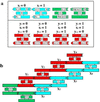



References
-
- Seeman N C. Trends Biotechnol. 1999;17:437–443. - PubMed
-
- Seeman N C. J Theor Biol. 1982;99:237–247. - PubMed
-
- Qiu H, Dewan J C, Seeman N C. J Mol Biol. 1997;267:881–898. - PubMed
-
- Robinson B H, Seeman N C. Protein Eng. 1987;1:295–300. - PubMed
-
- Petrillo M L, Newton C J, Cunningham R P, Ma R-I, Kallenbach N R, Seeman N C. Biopolymers. 1988;27:1337–1352. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

