Quantitative-trait homozygosity and association mapping and empirical genomewide significance in large, complex pedigrees: fasting serum-insulin level in the Hutterites
- PMID: 11880950
- PMCID: PMC379120
- DOI: 10.1086/339705
Quantitative-trait homozygosity and association mapping and empirical genomewide significance in large, complex pedigrees: fasting serum-insulin level in the Hutterites
Abstract
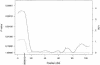
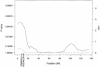
We present methods for linkage and association mapping of quantitative traits for a founder population with a large, known genealogy. We detect linkage to quantitative-trait loci (QTLs) through a multipoint homozygosity-mapping method. We propose two association methods, one of which is single point and uses a general two-allele model and the other of which is multipoint and uses homozygosity by descent for a particular allele. In all three methods, we make extensive use of the pedigree and genotype information, while keeping the computations simple and efficient. To assess significance, we have developed a permutation-based test that takes into account the covariance structure due to relatedness of individuals and can be used to determine empirical genomewide and locus-specific P values. In the case of multivariate-normally distributed trait data, the permutation-based test is asymptotically exact. The test is broadly applicable to a variety of mapping methods that fall within the class of linear statistical models (e.g., variance-component methods), under the assumption of random ascertainment with respect to the phenotype. For obtaining genomewide P values, our proposed method is appropriate when positions of markers are independent of the observed linkage signal, under the null hypothesis. We apply our methods to a genome screen for fasting insulin level in the Hutterites. We detect significant genomewide linkage on chromosome 19 and suggestive evidence of QTLs on chromosomes 1 and 16.
Figures



References
Electronic-Database Information
-
- Center for Medical Genetics, Marshfield Medical Research Foundation, http://research.marshfieldclinic.org/genetics/ (for microsatellite map)
References
-
- Anderson MJ, Robinson J (2001) Permutation tests for linear models. Aust NZ J Stat 43:75–88
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

