Coding sequences upstream of the human immunodeficiency virus type 1 reverse transcriptase domain in Gag-Pol are not essential for incorporation of the Pr160(gag-pol) into virus particles
- PMID: 11884546
- PMCID: PMC136043
- DOI: 10.1128/jvi.76.7.3221-3231.2002
Coding sequences upstream of the human immunodeficiency virus type 1 reverse transcriptase domain in Gag-Pol are not essential for incorporation of the Pr160(gag-pol) into virus particles
Abstract
Incorporation of the human immunodeficiency virus type 1 (HIV-1) Gag-Pol into virions is thought to be mediated by the N-terminal Gag domain via interaction with the Gag precursor. However, one recent study has demonstrated that the murine leukemia virus Pol can be incorporated into virions independently of Gag-Pol expression, implying a possible interaction between the Pol and Gag precursor. To test whether the HIV-1 Pol can be incorporated into virions on removal of the N-terminal Gag domain and to define sequences required for the incorporation of Gag-Pol into virions in more detail, a series of HIV Gag-Pol expression plasmids with various extensive deletions in the region upstream of the reverse transcriptase (RT) domain was constructed, and viral incorporation of the Gag-Pol deletion mutants was examined by cotransfecting 293T cells with a plasmid expressing Pr55(gag). Analysis indicated that deletion of the N-terminal two-thirds of the gag coding region did not significantly affect the incorporation of Gag-Pol into virions. In contrast, Gag-Pol proteins with deletions covering the capsid (CA) major homology regions and the adjacent C-terminal CA regions were impaired with respect to assembly into virions. However, Gag-Pol with sequences deleted upstream of the protease, or of the RT domain but retaining 15 N-terminal gag codons, could still be rescued into virions at a level about 20% of the wild-type level. When assayed in a nonmyristylated Gag-Pol context, all of the Gag-Pol deletion mutants were incorporated into virions at a level comparable to their myristylated counterparts, suggesting that the incorporation of the Gag-Pol deletion mutants into virions is independent of the N-terminal myristylation signal.
Figures




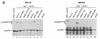
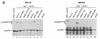
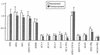
Similar articles
-
Characterization of human immunodeficiency virus type 1 Pr160 gag-pol mutants with truncations downstream of the protease domain.Virology. 2004 Nov 10;329(1):180-8. doi: 10.1016/j.virol.2004.08.010. Virology. 2004. PMID: 15476885
-
A domain directly C-terminal to the major homology region of human immunodeficiency type 1 capsid protein plays a crucial role in directing both virus assembly and incorporation of Gag-Pol.Virology. 2006 Apr 25;348(1):84-95. doi: 10.1016/j.virol.2005.12.009. Epub 2006 Jan 25. Virology. 2006. PMID: 16442581
-
Functional RT and IN incorporated into HIV-1 particles independently of the Gag/Pol precursor protein.EMBO J. 1997 Aug 15;16(16):5113-22. doi: 10.1093/emboj/16.16.5113. EMBO J. 1997. PMID: 9305652 Free PMC article.
-
Foamy virus assembly with emphasis on pol encapsidation.Viruses. 2013 Mar 20;5(3):886-900. doi: 10.3390/v5030886. Viruses. 2013. PMID: 23518575 Free PMC article. Review.
-
The packaging and maturation of the HIV-1 Pol proteins.Curr HIV Res. 2005 Jan;3(1):73-85. doi: 10.2174/1570162052772942. Curr HIV Res. 2005. PMID: 15638725 Review.
Cited by
-
HIV-1 Mutant Assembly, Processing and Infectivity Expresses Pol Independent of Gag.Viruses. 2020 Jan 2;12(1):54. doi: 10.3390/v12010054. Viruses. 2020. PMID: 31906562 Free PMC article.
-
Gag-Pol Transframe Domain p6* Is Essential for HIV-1 Protease-Mediated Virus Maturation.PLoS One. 2015 Jun 1;10(6):e0127974. doi: 10.1371/journal.pone.0127974. eCollection 2015. PLoS One. 2015. PMID: 26030443 Free PMC article.
-
Rapid localization of Gag/GagPol complexes to detergent-resistant membrane during the assembly of human immunodeficiency virus type 1.J Virol. 2003 Apr;77(7):3973-84. doi: 10.1128/jvi.77.7.3973-3984.2003. J Virol. 2003. PMID: 12634357 Free PMC article.
-
Sequential deletion of the integrase (Gag-Pol) carboxyl terminus reveals distinct phenotypic classes of defective HIV-1.J Virol. 2011 May;85(10):4654-66. doi: 10.1128/JVI.02374-10. Epub 2011 Mar 2. J Virol. 2011. PMID: 21367893 Free PMC article.
-
Placement of leucine zipper motifs at the carboxyl terminus of HIV-1 protease significantly reduces virion production.PLoS One. 2012;7(3):e32845. doi: 10.1371/journal.pone.0032845. Epub 2012 Mar 1. PLoS One. 2012. PMID: 22396796 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

