Distribution of spontaneous mutants and inferences about the replication mode of the RNA bacteriophage phi6
- PMID: 11884552
- PMCID: PMC136006
- DOI: 10.1128/jvi.76.7.3276-3281.2002
Distribution of spontaneous mutants and inferences about the replication mode of the RNA bacteriophage phi6
Abstract
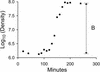
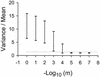
When a parent virus replicates inside its host, it must first use its own genome as the template for replication. However, once progeny genomes are produced, the progeny can in turn act as templates. Depending on whether the progeny genomes become templates, the distribution of mutants produced by an infection varies greatly. While information on the distribution is important for many population genetic models, it is also useful for inferring the replication mode of a virus. We have analyzed the distribution of mutants emerging from single bursts in the RNA bacteriophage phi6 and find that the distribution closely matches a Poisson distribution. The match suggests that replication in this bacteriophage is effectively by a stamping machine model in which the parental genome is the main template used for replication. However, because the distribution deviates slightly from a Poisson distribution, the stamping machine is not perfect and some progeny genomes must replicate. By fitting our data to a replication model in which the progeny genomes become replicative at a given rate or probability per round of replication, we estimated the rate to be very low and on the on the order of 10(-4). We discuss whether different replication modes may confer an adaptive advantage to viruses.
Figures
Similar articles
-
Packaging, replication and recombination of the segmented genome of bacteriophage Phi6 and its relatives.Virus Res. 2004 Apr;101(1):83-92. doi: 10.1016/j.virusres.2003.12.008. Virus Res. 2004. PMID: 15010219 Review.
-
How does the genome structure and lifestyle of a virus affect its population variation?Curr Opin Virol. 2014 Dec;9:39-44. doi: 10.1016/j.coviro.2014.09.004. Epub 2014 Sep 20. Curr Opin Virol. 2014. PMID: 25243801 Review.
-
The advantage of sex in the RNA virus phi6.Genetics. 1997 Nov;147(3):953-9. doi: 10.1093/genetics/147.3.953. Genetics. 1997. PMID: 9383044 Free PMC article.
-
Evolvability of an RNA virus is determined by its mutational neighbourhood.Nature. 2000 Aug 10;406(6796):625-8. doi: 10.1038/35020564. Nature. 2000. PMID: 10949302
-
Hybrid frequencies confirm limit to coinfection in the RNA bacteriophage phi6.J Virol. 1999 Mar;73(3):2420-4. doi: 10.1128/JVI.73.3.2420-2424.1999. J Virol. 1999. PMID: 9971826 Free PMC article.
Cited by
-
Does mutational robustness inhibit extinction by lethal mutagenesis in viral populations?PLoS Comput Biol. 2010 Jun 10;6(6):e1000811. doi: 10.1371/journal.pcbi.1000811. PLoS Comput Biol. 2010. PMID: 20548958 Free PMC article.
-
Emergence of Southern Rice Black-Streaked Dwarf Virus in the Centuries-Old Chinese Yuanyang Agrosystem of Rice Landraces.Viruses. 2019 Oct 25;11(11):985. doi: 10.3390/v11110985. Viruses. 2019. PMID: 31731529 Free PMC article.
-
Single-cell mutation rate of turnip crinkle virus (-)-strand replication intermediates.PLoS Pathog. 2023 Aug 14;19(8):e1011395. doi: 10.1371/journal.ppat.1011395. eCollection 2023 Aug. PLoS Pathog. 2023. PMID: 37578959 Free PMC article.
-
Variation in RNA virus mutation rates across host cells.PLoS Pathog. 2014 Jan;10(1):e1003855. doi: 10.1371/journal.ppat.1003855. Epub 2014 Jan 23. PLoS Pathog. 2014. PMID: 24465205 Free PMC article.
-
Genetically Determined Variation in Lysis Time Variance in the Bacteriophage φX174.G3 (Bethesda). 2016 Apr 7;6(4):939-55. doi: 10.1534/g3.115.024075. G3 (Bethesda). 2016. PMID: 26921293 Free PMC article.
References
-
- Burch, L. C., and L. Chao. 2000. Evolvability of an RNA virus determined by its mutational neighborhood. Nature 406:625-628. - PubMed
-
- Chao, L. 1991. Levels of selection, evolution of sex in RNA viruses, and the origin of life. J. Theor. Biol. 153:229-246. - PubMed
-
- Chao, L. 1990. Fitness of RNA virus decreased by Muller's ratchet. Nature 348:454-455. - PubMed
-
- Chao, L. 1992. Evolution of sex in RNA viruses. Trends Ecol. Evol. 7:147-151. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

