Nuclear interactions are necessary for translational enhancement by spleen necrosis virus RU5
- PMID: 11884554
- PMCID: PMC136029
- DOI: 10.1128/jvi.76.7.3292-3300.2002
Nuclear interactions are necessary for translational enhancement by spleen necrosis virus RU5
Abstract
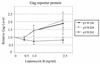
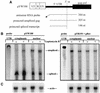
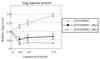
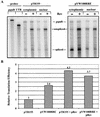
The 5' long terminal repeat of spleen necrosis virus (SNV) facilitates Rev/Rev-responsive element (RRE)-independent expression of intron-containing human immunodeficiency virus type 1 (HIV-1) gag. The SNV RU5 region, which corresponds to the 165-nucleotide 5' RNA terminus, functions in a position- and orientation-dependent manner to enhance polysome association of intron-containing HIV-1 gag RNA and also nonviral luc RNA. Evidence is mounting that association with nuclear factors during intron removal licenses mRNAs for nuclear export, efficient translation, and nonsense-mediated decay. This project addressed the relationship between the nuclear export pathway of SNV RU5-reporter RNA and translational enhancement. Results of RNA transfection experiments suggest that cytoplasmic proteins are insufficient for SNV RU5 translational enhancement of gag or luc RNA. Reporter gene assays, leptomycin B (LMB) sensitivity experiments, and RNase protection assays indicate that RU5 gag RNA accesses a nuclear export pathway that is distinct from the LMB-inhibited leucine-rich nuclear export sequence-dependent CRM1 pathway, which is used by the HIV-1 RRE. As a unique tool with which to investigate the relationship between different RNA trafficking routes and translational enhancement, SNV RU5 and Rev/RRE were combined on a single gag RNA. We observed a less-than-synergistic effect on cytoplasmic mRNA utilization. Instead, Rev/RRE diverts RU5 gag RNA to the CRM1-dependent, LMB-inhibited pathway and abrogates translational enhancement by SNV RU5. Our study is the first to show that a nuclear factor(s) directs SNV RU5-containing RNAs to a distinct export pathway that is not inhibited by LMB and programs the intron-containing transcript for translational enhancement.
Figures


Similar articles
-
The 5' RNA terminus of spleen necrosis virus stimulates translation of nonviral mRNA.J Virol. 2000 Sep;74(17):8111-8. doi: 10.1128/jvi.74.17.8111-8118.2000. J Virol. 2000. PMID: 10933721 Free PMC article.
-
Primary sequence and secondary structure motifs in spleen necrosis virus RU5 confer translational utilization of unspliced human immunodeficiency virus type 1 reporter RNA.J Virol. 2003 Nov;77(22):11973-84. doi: 10.1128/jvi.77.22.11973-11984.2003. J Virol. 2003. PMID: 14581534 Free PMC article.
-
The 5' RNA terminus of spleen necrosis virus contains a novel posttranscriptional control element that facilitates human immunodeficiency virus Rev/RRE-independent Gag production.J Virol. 1999 Jun;73(6):4847-55. doi: 10.1128/JVI.73.6.4847-4855.1999. J Virol. 1999. PMID: 10233946 Free PMC article.
-
Retroviral RNA elements integrate components of post-transcriptional gene expression.Life Sci. 2001 Oct 26;69(23):2697-709. doi: 10.1016/s0024-3205(01)01360-1. Life Sci. 2001. PMID: 11720075 Review.
-
Nucleocytoplasmic RNA transport in retroviral replication.Results Probl Cell Differ. 2001;34:197-217. doi: 10.1007/978-3-540-40025-7_12. Results Probl Cell Differ. 2001. PMID: 11288676 Review.
Cited by
-
Pericentriolar Targeting of the Mouse Mammary Tumor Virus GAG Protein.PLoS One. 2015 Jun 29;10(6):e0131515. doi: 10.1371/journal.pone.0131515. eCollection 2015. PLoS One. 2015. PMID: 26121257 Free PMC article.
-
The U3 region of Moloney murine leukemia virus contains position-independent cis-acting sequences involved in the nuclear export of full-length viral transcripts.J Biol Chem. 2014 Jul 18;289(29):20158-69. doi: 10.1074/jbc.M113.545855. Epub 2014 May 30. J Biol Chem. 2014. PMID: 24878957 Free PMC article.
-
Rev and Rex proteins of human complex retroviruses function with the MMTV Rem-responsive element.Retrovirology. 2009 Feb 3;6:10. doi: 10.1186/1742-4690-6-10. Retrovirology. 2009. PMID: 19192308 Free PMC article.
-
Gammaretrovirus mRNA expression is mediated by a novel, bipartite post-transcriptional regulatory element.Nucleic Acids Res. 2014;42(17):11092-106. doi: 10.1093/nar/gku798. Epub 2014 Sep 4. Nucleic Acids Res. 2014. PMID: 25190459 Free PMC article.
-
Analysis of synergy between divergent simple retrovirus posttranscriptional control elements.Virology. 2003 Dec 5;317(1):146-54. doi: 10.1016/j.virol.2003.08.037. Virology. 2003. PMID: 14675633 Free PMC article.
References
-
- Arrigo, S. J., and I. S. Chen. 1991. Rev is necessary for translation but not cytoplasmic accumulation of HIV-1 vif, vpr, and env/vpu 2 RNAs. Genes Dev. 5:808-819. - PubMed
-
- Banks, J. D., K. L. Beemon, and M. L. Linial. 1998. RNA regulatory elements in the genomes of simple retroviruses. Semin. Virol. 8:194-204.
-
- Boris-Lawrie, K., T. M. Roberts, and S. Hull. 2001. Retroviral RNA elements integrate components of post-transcriptional gene expression. Life Sci. 69:2697-2709. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

