Replication of bovine papillomavirus type 1 (BPV-1) DNA in Saccharomyces cerevisiae following infection with BPV-1 virions
- PMID: 11884561
- PMCID: PMC136048
- DOI: 10.1128/jvi.76.7.3359-3364.2002
Replication of bovine papillomavirus type 1 (BPV-1) DNA in Saccharomyces cerevisiae following infection with BPV-1 virions
Abstract
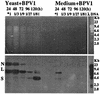
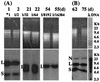
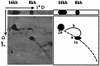
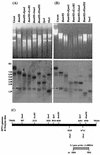
Saccharomyces cerevisiae protoplasts exposed to bovine papillomavirus type 1 (BPV-1) virions demonstrated uptake of virions on electron microscopy. S. cerevisiae cells looked larger after exposure to BPV-1 virions, and cell wall regeneration was delayed. Southern blot hybridization of Hirt DNA from cells exposed to BPV-1 virions demonstrated BPV-1 DNA, which could be detected over 80 days of culture and at least 13 rounds of division. Two-dimensional gel analysis of Hirt DNA showed replicative intermediates, confirming that the BPV-1 genome was replicating within S. cerevisiae. Nicked circle, linear, and supercoiled BPV-1 DNA species were observed in Hirt DNA preparations from S. cerevisiae cells infected for over 50 days, and restriction digestion showed fragments hybridizing to BPV-1 in accord with the predicted restriction map for circular BPV-1 episomes. These data suggest that BPV-1 can infect S. cerevisiae and that BPV-1 episomes can replicate in the infected S. cerevisiae cells.
Figures

Similar articles
-
Characterization of Episomal Replication of Bovine Papillomavirus Type 1 DNA in Long-Term Virion-Infected Saccharomyces Cerevisiae Culture.Virol Sin. 2021 Dec;36(6):1492-1502. doi: 10.1007/s12250-021-00439-y. Epub 2021 Aug 30. Virol Sin. 2021. PMID: 34460066 Free PMC article.
-
Saccharomyces cerevisiae is permissive for replication of bovine papillomavirus type 1.J Virol. 2002 Dec;76(23):12265-73. doi: 10.1128/jvi.76.23.12265-12273.2002. J Virol. 2002. PMID: 12414966 Free PMC article.
-
Rolling-circle replication of a high-copy BPV-1 plasmid.J Mol Biol. 1992 Nov 5;228(1):1-6. doi: 10.1016/0022-2836(92)90485-3. J Mol Biol. 1992. PMID: 1333015
-
Replication of plasmids derived from bovine papilloma virus type 1 and Epstein-Barr virus in cells in culture.Annu Rev Cell Biol. 1987;3:87-108. doi: 10.1146/annurev.cb.03.110187.000511. Annu Rev Cell Biol. 1987. PMID: 2825737 Review.
-
Saccharomyces cerevisiae: a useful model host to study fundamental biology of viral replication.Virus Res. 2006 Sep;120(1-2):49-56. doi: 10.1016/j.virusres.2005.11.018. Epub 2006 May 15. Virus Res. 2006. PMID: 16698107 Free PMC article. Review.
Cited by
-
Replication and encapsidation of papillomaviruses in Saccharomyces cerevisiae.Methods Mol Med. 2005;119:247-60. doi: 10.1385/1-59259-982-6:247. Methods Mol Med. 2005. PMID: 16353338 Free PMC article.
-
Characterization of Episomal Replication of Bovine Papillomavirus Type 1 DNA in Long-Term Virion-Infected Saccharomyces Cerevisiae Culture.Virol Sin. 2021 Dec;36(6):1492-1502. doi: 10.1007/s12250-021-00439-y. Epub 2021 Aug 30. Virol Sin. 2021. PMID: 34460066 Free PMC article.
-
Modeling the function of bacterial virulence factors in Saccharomyces cerevisiae.Eukaryot Cell. 2004 Aug;3(4):827-34. doi: 10.1128/EC.3.4.827-834.2004. Eukaryot Cell. 2004. PMID: 15302815 Free PMC article. Review. No abstract available.
-
Saccharomyces cerevisiae: a versatile eukaryotic system in virology.Microb Cell Fact. 2007 Oct 10;6:32. doi: 10.1186/1475-2859-6-32. Microb Cell Fact. 2007. PMID: 17927824 Free PMC article.
-
Stable replication of papillomavirus genomes in Saccharomyces cerevisiae.J Virol. 2002 Apr;76(7):3350-8. doi: 10.1128/jvi.76.7.3350-3358.2002. J Virol. 2002. PMID: 11884560 Free PMC article.
References
-
- Bendel, C. M., J. St Sauver, S. Carlson, and M. K. Hostetter. 1995. Epithelial adhesion in yeast species: correlation with surface expression of the integrin analog. J. Infect. Dis. 171:1660-1663. - PubMed
-
- Cheng, S., D. C. Schmidt-Grimminger, T. Murant, T. R. Broker, and L. T. Chow. 1995. Differentiation-dependent up-regulation of the human papillomavirus E7 gene reactivates cellular DNA replication in suprabasal differentiated keratinocytes. Genes Dev. 9:2335-2349. - PubMed
-
- Dollard, S. C., J. L. Wilson, L. M. Demeter, W. Bonnez, R. C. Reichman, T. R. Broker, and L. T. Chow. 1992. Production of human papillomavirus and modulation of the infectious program in epithelial raft cultures. Genes Dev. 6:1131-1142. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

