Evolutionary history of Cucumber mosaic virus deduced by phylogenetic analyses
- PMID: 11884564
- PMCID: PMC136033
- DOI: 10.1128/jvi.76.7.3382-3387.2002
Evolutionary history of Cucumber mosaic virus deduced by phylogenetic analyses
Abstract
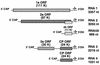
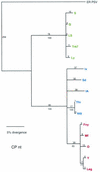
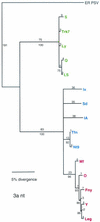
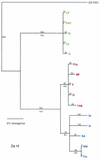
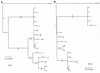
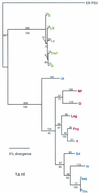
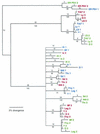
Cucumber mosaic virus (CMV) is an RNA plant virus with a tripartite genome and an extremely broad host range. Previous evolutionary analyses with the coat protein (CP) and 5' nontranslated region (NTR) of RNA 3 suggested subdivision of the virus into three groups, subgroups IA, IB, and II. In this study 15 strains of CMV whose nucleotide sequences have been determined were used for a complete phylogenetic analysis of the virus. The trees estimated for open reading frames (ORFs) located on the different RNAs were not congruent and did not completely support the subgrouping indicated by the CP ORF, indicating that different RNAs had independent evolutionary histories. This is consistent with a reassortment mechanism playing an important role in the evolution of the virus. The evolutionary trees of the 1a and 3a ORFs were more compact and displayed more branching than did those of the 2a and CP ORFs. This may reflect more rigid host-interactive constraints exerted on the 1a and 3a ORFs. In addition, analysis of the 3' NTR that is conserved among all RNAs indicated that evolutionary constraints on this region are specific to the RNA component rather than the virus isolate. This indicates that functions other than replication are encoded in the 3' NTR. Reassortment may have led to the genetic diversity found among CMV strains and contributed to its enormous evolutionary success.
Figures
Similar articles
-
Role of recombination in the evolution of natural populations of Cucumber mosaic virus, a tripartite RNA plant virus.Virology. 2005 Feb 5;332(1):359-68. doi: 10.1016/j.virol.2004.11.017. Virology. 2005. PMID: 15661167
-
Rearrangements in the 5' nontranslated region and phylogenetic analyses of cucumber mosaic virus RNA 3 indicate radial evolution of three subgroups.J Virol. 1999 Aug;73(8):6752-8. doi: 10.1128/JVI.73.8.6752-6758.1999. J Virol. 1999. PMID: 10400773 Free PMC article.
-
Molecular evidence and sequence analysis of a natural reassortant between cucumber mosaic virus subgroup IA and II strains.Virus Genes. 2007 Oct;35(2):405-13. doi: 10.1007/s11262-007-0094-z. Epub 2007 Apr 7. Virus Genes. 2007. PMID: 17417698
-
Cucumber mosaic virus: viral genes as virulence determinants.Mol Plant Pathol. 2012 Apr;13(3):217-25. doi: 10.1111/j.1364-3703.2011.00749.x. Epub 2011 Oct 7. Mol Plant Pathol. 2012. PMID: 21980997 Free PMC article. Review.
-
RNA Virus Reassortment: An Evolutionary Mechanism for Host Jumps and Immune Evasion.PLoS Pathog. 2015 Jul 9;11(7):e1004902. doi: 10.1371/journal.ppat.1004902. eCollection 2015 Jul. PLoS Pathog. 2015. PMID: 26158697 Free PMC article. Review. No abstract available.
Cited by
-
Fixation of emerging interviral recombinants in cucumber mosaic virus populations.J Virol. 2013 Jan;87(2):1264-9. doi: 10.1128/JVI.01892-12. Epub 2012 Oct 31. J Virol. 2013. PMID: 23115282 Free PMC article.
-
Ethylene emitted by viral pathogen-infected pepper (Capsicum annuum L.) plants is a volatile chemical cue that attracts aphid vectors.Front Plant Sci. 2022 Sep 29;13:994314. doi: 10.3389/fpls.2022.994314. eCollection 2022. Front Plant Sci. 2022. PMID: 36247604 Free PMC article.
-
Pseudorecombination between Two Distinct Strains of Cucumber mosaic virus Results in Enhancement of Symptom Severity.Plant Pathol J. 2014 Sep;30(3):316-22. doi: 10.5423/PPJ.NT.04.2014.0031. Plant Pathol J. 2014. PMID: 25289019 Free PMC article.
-
Evolutionary ecology of virus emergence.Ann N Y Acad Sci. 2017 Feb;1389(1):124-146. doi: 10.1111/nyas.13304. Epub 2016 Dec 30. Ann N Y Acad Sci. 2017. PMID: 28036113 Free PMC article. Review.
-
Genetic Structure of Cucumber Mosaic Virus From Natural Hosts in Nigeria Reveals High Diversity and Occurrence of Putative Novel Recombinant Strains.Front Microbiol. 2022 Feb 10;13:753054. doi: 10.3389/fmicb.2022.753054. eCollection 2022. Front Microbiol. 2022. PMID: 35222322 Free PMC article.
References
-
- Chao, L., T. Tran, and C. Matthews. 1992. Muller's ratchet and the advantage of sex in the RNA virus ø6. Evolution 46:289-299. - PubMed
-
- Ding, S.-W., B. J. Anderson, H. R. Haase, and R. H. Symons. 1994. New overlapping gene encoded by the cucumber mosaic virus genome. Virology 198:593-601. - PubMed
-
- Doolittle, S. P. 1916. A new infectious mosaic disease of cucumber. Phytopathology 6:145-147.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

