The PHD type zinc finger is an integral part of the CBP acetyltransferase domain
- PMID: 11884585
- PMCID: PMC133676
- DOI: 10.1128/MCB.22.7.1961-1970.2002
The PHD type zinc finger is an integral part of the CBP acetyltransferase domain
Abstract
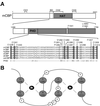
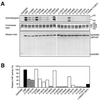
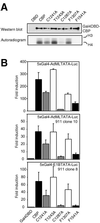
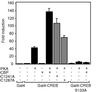
Histone acetyltransferases (HATs) such as CBP and p300 are regarded as key regulators of RNA polymerase II-mediated transcription, but the critical structural features of their HAT modules remain ill defined. The HAT domains of CBP and p300 are characterized by the presence of a highly conserved putative plant homeodomain (PHD) (C4HC3) type zinc finger, which is part of the functionally uncharacterized cysteine-histidine-rich region 2 (CH2). Here we show that this region conforms to the PHD type zinc finger consensus and that it is essential for in vitro acetylation of core histones and the basal transcription factor TFIIE34 as well as for CBP autoacetylation. PHD finger mutations also reduced the transcriptional activity of the full-length CBP protein when tested on transfected reporter genes. Importantly, similar results were obtained on integrated reporters, which reflect a more natural chromatinized state. Taken together, our results indicate that the PHD finger forms an integral part of the enzymatic core of the HAT domain of CBP.
Figures




Similar articles
-
Functional analysis of the p300 acetyltransferase domain: the PHD finger of p300 but not of CBP is dispensable for enzymatic activity.Nucleic Acids Res. 2001 Nov 1;29(21):4462-71. doi: 10.1093/nar/29.21.4462. Nucleic Acids Res. 2001. PMID: 11691934 Free PMC article.
-
Acetyltransferase machinery conserved in p300/CBP-family proteins.Oncogene. 2002 Mar 28;21(14):2253-60. doi: 10.1038/sj.onc.1205283. Oncogene. 2002. PMID: 11948408
-
Stimulation of CREB binding protein nucleosomal histone acetyltransferase activity by a class of transcriptional activators.Mol Cell Biol. 2001 Jan;21(2):476-87. doi: 10.1128/MCB.21.2.476-487.2001. Mol Cell Biol. 2001. PMID: 11134336 Free PMC article.
-
CBP and p300: HATs for different occasions.Biochem Pharmacol. 2004 Sep 15;68(6):1145-55. doi: 10.1016/j.bcp.2004.03.045. Biochem Pharmacol. 2004. PMID: 15313412 Review.
-
CBP, a transcriptional coactivator and acetyltransferase.Biochem Cell Biol. 2001;79(3):253-66. Biochem Cell Biol. 2001. PMID: 11467739 Review.
Cited by
-
Structural insight into ASH1L PHD finger recognizing methylated histone H3K4 and promoting cell growth in prostate cancer.Front Oncol. 2022 Aug 10;12:906807. doi: 10.3389/fonc.2022.906807. eCollection 2022. Front Oncol. 2022. PMID: 36033518 Free PMC article.
-
A plant homeodomain in RAG-2 that binds Hypermethylated lysine 4 of histone H3 is necessary for efficient antigen-receptor-gene rearrangement.Immunity. 2007 Oct;27(4):561-71. doi: 10.1016/j.immuni.2007.09.005. Epub 2007 Oct 11. Immunity. 2007. PMID: 17936034 Free PMC article.
-
Soybean GmPHD-type transcription regulators improve stress tolerance in transgenic Arabidopsis plants.PLoS One. 2009 Sep 30;4(9):e7209. doi: 10.1371/journal.pone.0007209. PLoS One. 2009. PMID: 19789627 Free PMC article.
-
Lint-O cooperates with L(3)mbt in target gene suppression to maintain homeostasis in fly ovary and brain.EMBO Rep. 2022 Oct 6;23(10):e53813. doi: 10.15252/embr.202153813. Epub 2022 Aug 22. EMBO Rep. 2022. PMID: 35993198 Free PMC article.
-
Zinc Finger 280B regulates sGCα1 and p53 in prostate cancer cells.PLoS One. 2013 Nov 13;8(11):e78766. doi: 10.1371/journal.pone.0078766. eCollection 2013. PLoS One. 2013. PMID: 24236047 Free PMC article.
References
-
- Aasland, R., T. J. Gibson, and A. F. Stewart. 1995. The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem. Sci. 20:56-59. - PubMed
-
- Ait-Si-Ali, S., S. Ramirez, F. X. Barre, F. Dkhissi, L. Magnaghi-Jaulin, J. A. Girault, P. Robin, M. Knibiehler, L. L. Pritchard, B. Ducommun, D. Trouche, and A. Harel-Bellan. 1998. Histone acetyltransferase activity of CBP is controlled by cycle-dependent kinases and oncoprotein E1A. Nature 396:184-186. - PubMed
-
- Arany, Z., W. R. Sellers, D. M. Livingston, and R. Eckner. 1994. E1A-associated p300 and CREB-associated CBP belong to a conserved family of coactivators. Cell 77:799-800. - PubMed
-
- Arias, J., A. S. Alberts, P. Brindle, F. X. Claret, T. Smeal, M. Karin, J. Feramisco, and M. Montminy. 1994. Activation of cAMP and mitogen responsive genes relies on a common nuclear factor. Nature 370:226-229. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
