Ime2, a meiosis-specific kinase in yeast, is required for destabilization of its transcriptional activator, Ime1
- PMID: 11884593
- PMCID: PMC133691
- DOI: 10.1128/MCB.22.7.2047-2056.2002
Ime2, a meiosis-specific kinase in yeast, is required for destabilization of its transcriptional activator, Ime1
Abstract
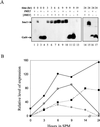
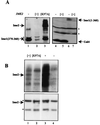
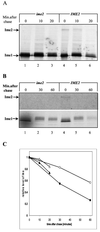
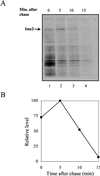
In the budding yeast Saccharomyces cerevisiae, entry into meiosis and its successful completion depend on two positive regulators, Ime1 and Ime2. Ime1 is a transcriptional activator that is required for transcription of IME2, a serine/threonine protein kinase. We show that in vivo Ime2 associates with Ime1, that in vitro Ime2 phosphorylates Ime1, and that in living cells the stability of Ime1 depends on Ime2. Diploid cells with IME2 deleted show an increase in the level of Ime1, whereas haploid cells overexpressing IME2 show a decrease in the stability of Ime1. Furthermore, the level of Ime1 depends on the kinase activity of Ime2. Using a mutation in one of the ATPase subunits of the proteasome, RPT2, we demonstrate that Ime1, amino acids 270 to 360, is degraded by the 26S proteasome. We also show that Ime2 itself is an extremely unstable protein whose expression in vegetative cultures is toxic. We propose that a negative-feedback loop ensures that the activity of Ime1 will be restricted to a narrow window.
Figures




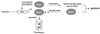
References
-
- Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. - PubMed
-
- Chu, S., J. DeRisi, M. Eisen, J. Mulholland, D. Botstein, P. O. Brown, and I. Herskowitz. 1998. The transcriptional program of sporulation in budding yeast. Science 282:699-705. (Erratum, 282:1421.) - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
