Targeted transposition by the V(D)J recombinase
- PMID: 11884595
- PMCID: PMC133684
- DOI: 10.1128/MCB.22.7.2068-2077.2002
Targeted transposition by the V(D)J recombinase
Abstract
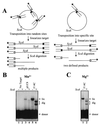
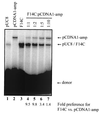
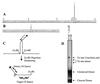
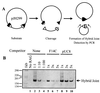
Cleavage by the V(D)J recombinase at a pair of recombination signal sequences creates two coding ends and two signal ends. The RAG proteins can integrate these signal ends, without sequence specificity, into an unrelated target DNA molecule. Here we demonstrate that such transposition events are greatly stimulated by--and specifically targeted to--hairpins and other distorted DNA structures. The mechanism of target selection by the RAG proteins thus appears to involve recognition of distorted DNA. These data also suggest a novel mechanism for the formation of alternative recombination products termed hybrid joints, in which a signal end is joined to a hairpin coding end. We suggest that hybrid joints may arise by transposition in vivo and propose a new model to account for some recurrent chromosome translocations found in human lymphomas. According to this model, transposition can join antigen receptor loci to partner sites that lack recombination signal sequence elements but bear particular structural features. The RAG proteins are capable of mediating all necessary breakage and joining events on both partner chromosomes; thus, the V(D)J recombinase may be far more culpable for oncogenic translocations than has been suspected.
Figures


Similar articles
-
In vivo transposition mediated by V(D)J recombinase in human T lymphocytes.EMBO J. 2003 Mar 17;22(6):1381-8. doi: 10.1093/emboj/cdg137. EMBO J. 2003. PMID: 12628930 Free PMC article.
-
The DDE motif in RAG-1 is contributed in trans to a single active site that catalyzes the nicking and transesterification steps of V(D)J recombination.Mol Cell Biol. 2001 Jan;21(2):449-58. doi: 10.1128/MCB.21.2.449-458.2001. Mol Cell Biol. 2001. PMID: 11134333 Free PMC article.
-
The V(D)J recombinase efficiently cleaves and transposes signal joints.Mol Cell. 2002 Apr;9(4):871-8. doi: 10.1016/s1097-2765(02)00494-x. Mol Cell. 2002. PMID: 11983177
-
The RAG proteins and V(D)J recombination: complexes, ends, and transposition.Annu Rev Immunol. 2000;18:495-527. doi: 10.1146/annurev.immunol.18.1.495. Annu Rev Immunol. 2000. PMID: 10837067 Review.
-
RAG1 and RAG2 in V(D)J recombination and transposition.Immunol Res. 2001;23(1):23-39. doi: 10.1385/IR:23:1:23. Immunol Res. 2001. PMID: 11417858 Review.
Cited by
-
In vivo transposition mediated by V(D)J recombinase in human T lymphocytes.EMBO J. 2003 Mar 17;22(6):1381-8. doi: 10.1093/emboj/cdg137. EMBO J. 2003. PMID: 12628930 Free PMC article.
-
Double-strand break formation by the RAG complex at the bcl-2 major breakpoint region and at other non-B DNA structures in vitro.Mol Cell Biol. 2005 Jul;25(14):5904-19. doi: 10.1128/MCB.25.14.5904-5919.2005. Mol Cell Biol. 2005. PMID: 15988007 Free PMC article.
-
In vivo reinsertion of excised episomes by the V(D)J recombinase: a potential threat to genomic stability.PLoS Biol. 2007 Mar;5(3):e43. doi: 10.1371/journal.pbio.0050043. PLoS Biol. 2007. PMID: 17298184 Free PMC article.
-
Monitoring single-stranded DNA secondary structure formation by determining the topological state of DNA catenanes.Biophys J. 2006 Apr 15;90(8):2877-89. doi: 10.1529/biophysj.105.074104. Epub 2006 Feb 3. Biophys J. 2006. PMID: 16461397 Free PMC article.
-
DNA mismatches and GC-rich motifs target transposition by the RAG1/RAG2 transposase.Nucleic Acids Res. 2003 Nov 1;31(21):6180-90. doi: 10.1093/nar/gkg819. Nucleic Acids Res. 2003. PMID: 14576304 Free PMC article.
References
-
- Agrawal, A., Q. M. Eastman, and D. G. Schatz. 1998. Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature 394:744-751. - PubMed
-
- Agrawal, A., and D. G. Schatz. 1997. RAG1 and RAG2 form a stable postcleavage synaptic complex with DNA containing signal ends in V(D)J recombination. Cell 89:43-53. - PubMed
-
- Besmer, E., J. Mansilia-Soto, S. Cassard, D. J. Sawchuk, G. Brown, M. Sadofsky, S. M. Lewis, M. C. Nussenzweig, and P. Cortes. 1998. Hairpin coding end opening is mediated by RAG1 and RAG2 proteins. Mol. Cell 2:817-828. - PubMed
-
- Bogue, M. A., C. Wang, C. Zhu, and D. B. Roth. 1997. V(D)J recombination in Ku86-deficient mice: distinct effects on coding, signal, and hybrid joint formation. Immunity 7:37-47. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
