Shared role for differentially methylated domains of imprinted genes
- PMID: 11884597
- PMCID: PMC133683
- DOI: 10.1128/MCB.22.7.2089-2098.2002
Shared role for differentially methylated domains of imprinted genes
Abstract
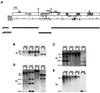
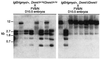
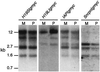
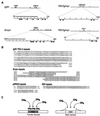
For most imprinted genes, a difference in expression between the maternal and paternal alleles is associated with a corresponding difference in DNA methylation that is localized to a differentially methylated domain (DMD). Removal of a gene's DMD leads to a loss of imprinting. These observations suggest that DMDs have a determinative role in genomic imprinting. To examine this possibility, we introduced sequences from the DMDs of the imprinted Igf2r, H19, and Snrpn genes into a nonimprinted derivative of the normally imprinted RSVIgmyc transgene, created by excising its own DMD. Hybrid transgenes with sequences from the Igf2r DMD2 were consistently imprinted, with the maternal allele being more methylated than the paternal allele. Only the repeated sequences within DMD2 were required for imprinting these transgenes. Hybrid transgenes containing H19 and Snrpn DMD sequences and ones containing sequences from the long terminal repeat of a murine intracisternal A particle retrotransposon were not imprinted. The Igf2r hybrid transgenes are comprised entirely of mouse genomic DNA and behave as endogenous imprinted genes in inbred wild-type and mutant mouse strains. These types of hybrid transgenes can be used to elucidate the functions of DMD sequences in genomic imprinting.
Figures



References
-
- Ainscough, J. F., T. Koide, M. Tada, S. Barton, and M. A. Surani. 1997. Imprinting of Igf2 and H19 from a 130 kb YAC transgene. Development 124:3621-3632. - PubMed
-
- Bartolomei, M. S., and S. M. Tilghman. 1997. Genomic imprinting in mammals. Annu. Rev. Genet. 31:493-525. - PubMed
-
- Bell, A. C., and G. Felsenfeld. 2000. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405:482-485. - PubMed
-
- Bielinska, B., S. M. Blaydes, K. Buiting, T. Yang, M. Krajewska-Walasek, B. Horsthemke, and C. I. Brannan. 2000. De novo deletions of SNRPN exon 1 in early human and mouse embryos result in a paternal to maternal imprint switch. Nat. Genet. 25:74-78. - PubMed
-
- Birger, Y., R. Shemer, J. Perk, and A. Razin. 1999. The imprinting box of the mouse Igf2r gene. Nature 397:84-88. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
