Reversal of growth suppression by p107 via direct phosphorylation by cyclin D1/cyclin-dependent kinase 4
- PMID: 11884610
- PMCID: PMC133692
- DOI: 10.1128/MCB.22.7.2242-2254.2002
Reversal of growth suppression by p107 via direct phosphorylation by cyclin D1/cyclin-dependent kinase 4
Abstract
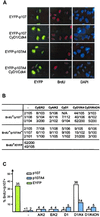
p107 functions to control cell division and development through interaction with members of the E2F family of transcription factors. p107 is phosphorylated in a cell cycle-regulated manner, and its phosphorylation leads to its release from E2F. Although it is known that p107 physically associates with E- and A-type cyclin/cyclin-dependent kinase 2 (Cdk2) complexes through a cyclin-binding RXL motif located in the spacer domain, the mechanisms underlying p107 inactivation via phosphorylation remain poorly defined. Recent genetic evidence indicates a requirement for cyclin D1/Cdk4 complexes in p107 inactivation. In this work, we provide direct biochemical evidence for the involvement of cyclin D1/Cdk4 in the inactivation of p107's growth-suppressive function. While coexpression of cyclin D1/Cdk4 can reverse the cell cycle arrest properties of p107 in Saos-2 cells, we find that p107 in which the Lys-Arg-Arg-Leu sequence of the RXL motif is replaced by four alanine residues is largely refractory to inactivation by cyclin D/Cdk4, indicating a role for this motif in p107 inactivation without a requirement for its tight interaction with cyclin D1/Cdk4. We identified four phosphorylation sites in p107 (Thr-369, Ser-640, Ser-964, and Ser-975) that are efficiently phosphorylated by Cdk4 but not by Cdk2 in vitro and are also phosphorylated in tissue culture cells. Growth suppression by p107 containing nonphosphorylatable residues in these four sites is not reversed by coexpression of cyclin D1/Cdk4. In model p107 spacer region peptides, phosphorylation of S640 by cyclin D1/Cdk4 is strictly dependent upon an intact RXL motif, but phosphorylation of this site in the absence of an RXL motif can be partially restored by replacement of S643 by arginine. This suggests that one role for the RXL motif is to facilitate phosphorylation of nonconsensus Cdk substrates. Taken together, these data indicate that p107 is inactivated by cyclin D1/Cdk4 via direct phosphorylation and that the RXL motif of p107 plays a role in its inactivation by Cdk4 in the absence of stable binding.
Figures








Similar articles
-
Progesterone inhibits estrogen-induced cyclin D1 and cdk4 nuclear translocation, cyclin E- and cyclin A-cdk2 kinase activation, and cell proliferation in uterine epithelial cells in mice.Mol Cell Biol. 1999 Mar;19(3):2251-64. doi: 10.1128/MCB.19.3.2251. Mol Cell Biol. 1999. PMID: 10022912 Free PMC article.
-
Expression and activity of the retinoblastoma protein (pRB)-family proteins, p107 and p130, during L6 myoblast differentiation.Cell Growth Differ. 1995 Oct;6(10):1287-98. Cell Growth Differ. 1995. PMID: 8845306
-
Disruption of the actin cytoskeleton leads to inhibition of mitogen-induced cyclin E expression, Cdk2 phosphorylation, and nuclear accumulation of the retinoblastoma protein-related p107 protein.Exp Cell Res. 2000 Aug 25;259(1):35-53. doi: 10.1006/excr.2000.4966. Exp Cell Res. 2000. PMID: 10942577
-
Cdk2-dependent phosphorylation and functional inactivation of the pRB-related p130 protein in pRB(-), p16INK4A(+) tumor cells.J Biol Chem. 2000 Sep 29;275(39):30317-25. doi: 10.1074/jbc.M005707200. J Biol Chem. 2000. PMID: 10906146
-
Cyclin B and cyclin A confer different substrate recognition properties on CDK2.Cell Cycle. 2007 Jun 1;6(11):1350-9. doi: 10.4161/cc.6.11.4278. Epub 2007 Jun 11. Cell Cycle. 2007. PMID: 17495531
Cited by
-
RB1, development, and cancer.Curr Top Dev Biol. 2011;94:129-69. doi: 10.1016/B978-0-12-380916-2.00005-X. Curr Top Dev Biol. 2011. PMID: 21295686 Free PMC article. Review.
-
Cyclin D3 deficiency inhibits skin tumor development, but does not affect normal keratinocyte proliferation.Oncol Lett. 2017 Sep;14(3):2723-2734. doi: 10.3892/ol.2017.6551. Epub 2017 Jul 8. Oncol Lett. 2017. PMID: 28927034 Free PMC article.
-
Deletion of the p107/p130-binding domain of Mip130/LIN-9 bypasses the requirement for CDK4 activity for the dissociation of Mip130/LIN-9 from p107/p130-E2F4 complex.Exp Cell Res. 2009 Oct 15;315(17):2914-20. doi: 10.1016/j.yexcr.2009.07.014. Epub 2009 Jul 18. Exp Cell Res. 2009. PMID: 19619530 Free PMC article.
-
Down-regulation of hTERT and Cyclin D1 transcription via PI3K/Akt and TGF-β pathways in MCF-7 Cancer cells with PX-866 and Raloxifene.Exp Cell Res. 2016 May 15;344(1):95-102. doi: 10.1016/j.yexcr.2016.03.022. Epub 2016 Mar 23. Exp Cell Res. 2016. PMID: 27017931 Free PMC article.
-
CDK1 structures reveal conserved and unique features of the essential cell cycle CDK.Nat Commun. 2015 Apr 13;6:6769. doi: 10.1038/ncomms7769. Nat Commun. 2015. PMID: 25864384 Free PMC article.
References
-
- Adams, P. D., and J. W. Harper. 2001. Cyclin-dependent kinases. Chem. Rev. 101:2511-2526. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
