Developmental control of histone mRNA and dSLBP synthesis during Drosophila embryogenesis and the role of dSLBP in histone mRNA 3' end processing in vivo
- PMID: 11884612
- PMCID: PMC133687
- DOI: 10.1128/MCB.22.7.2267-2282.2002
Developmental control of histone mRNA and dSLBP synthesis during Drosophila embryogenesis and the role of dSLBP in histone mRNA 3' end processing in vivo
Abstract
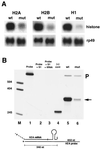
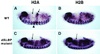
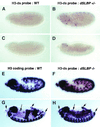
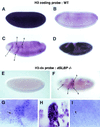
In metazoans, the 3' end of histone mRNA is not polyadenylated but instead ends with a stem-loop structure that is required for cell cycle-regulated expression. The sequence of the stem-loop in the Drosophila melanogaster histone H2b, H3, and H4 genes is identical to the consensus sequence of other metazoan histone mRNAs, but the sequence of the stem-loop in the D. melanogaster histone H2a and H1 genes is novel. dSLBP binds to these novel stem-loop sequences as well as the canonical stem-loop with similar affinity. Eggs derived from females containing a viable, hypomorphic mutation in dSLBP store greatly reduced amounts of all five histone mRNAs in the egg, indicating that dSLBP is required in the maternal germ line for production of each histone mRNA. Embryos deficient in zygotic dSLBP function accumulate poly(A)(+) versions of all five histone mRNAs as a result of usage of polyadenylation signals located 3' of the stem-loop in each histone gene. Since the 3' ends of adjacent histone genes are close together, these polyadenylation signals may ensure the termination of transcription in order to prevent read-through into the next gene, which could possibly disrupt transcription or produce antisense histone mRNA that might trigger RNA interference. During early wild-type embryogenesis, ubiquitous zygotic histone gene transcription is activated at the end of the syncytial nuclear cycles during S phase of cycle 14, silenced during the subsequent G(2) phase, and then reactivated near the end of that G(2) phase in the well-described mitotic domain pattern. There is little or no dSLBP protein provided maternally in wild-type embryos, and zygotic expression of dSLBP is immediately required to process newly made histone pre-mRNA.
Figures





References
-
- Akhmanova, A., H. Kremer, K. Miedema, and W. Hennig. 1997. Naturally occurring testis-specific histone H3 antisense transcripts in Drosophila. Mol. Reprod. Dev. 48:413-420. - PubMed
-
- Akhmanova, A., K. Miedema, H. Kremer, and W. Hennig. 1997. Two types of polyadenated mRNAs are synthesized from Drosophila replication-dependent histone genes. Eur J. Biochem. 244:294-300. - PubMed
-
- Calvo, O., and J. L. Manley. 2001. Evolutionarily conserved interaction between CstF-64 and PC4 links transcription, polyadenylation, and termination. Mol. Cell 7:1013-1023. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
