The solution structure of an oligonucleotide duplex containing a 2'-deoxyadenosine-3-(2-hydroxyethyl)- 2'-deoxyuridine base pair determined by NMR and molecular dynamics studies
- PMID: 11884635
- PMCID: PMC101359
- DOI: 10.1093/nar/30.6.1371
The solution structure of an oligonucleotide duplex containing a 2'-deoxyadenosine-3-(2-hydroxyethyl)- 2'-deoxyuridine base pair determined by NMR and molecular dynamics studies
Abstract
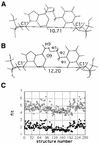
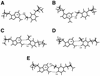
Determination of the solution structure of the duplex d(GCAAGTC(HE)AAAACG)*d(CGTTTTAGACTTGC) containing a 3-(2-hydroxyethyl)-2'-deoxyuridine*deoxyadenine (HE*A) base pair is reported. The three-dimensional solution structure, determined starting from 512 models via restrained molecular mechanics using inter-proton distances and torsion angles, converged to two final families of structures. For both families the HE and the opposite A residues are intrahelical and in the anti conformation. The hydroxyethyl chain lies close to the helix axis and for one family the hydroxyl group is above the HE*A plane and in the other case it is below. These two models were used to start molecular dynamic calculations with explicit solvent to explore the hydrogen bonding possibilities of the HE*A base pair. The dynamics calculations converge finally to one model structure in which two hydrogen bonds are formed. The first is formed all the time and is between HEO4 and the amino group of A, and the second, an intermittent one, is between the hydroxyl group and the N1 of A. When this second hydrogen bond is not formed a weak interaction CH...N is possible between HEC7H2 and N1A21. All the best structures show an increase in the C1'-C1' distance relative to a Watson-Crick base pair.
Figures





References
-
- Dellarco V.L., Generoso,W.M., Sega,G.A., Fowle,J.R.D. and Jacobson-Kram,D. (1990) Review of the mutagenicity of ethylene oxide. Environ. Mol. Mutagen., 16, 85–103. - PubMed
-
- Ehrenberg L. and Hussain,S. (1981) Genetic toxicity of some important epoxides. Mutat. Res., 86, 1–113. - PubMed
-
- Hogstedt C., Aringer,L. and Gustavsson,A. (1986) Epidemiologic support for ethylene oxide as a cancer-causing agent. J. Am. Med. Assoc., 255, 1575–1578. - PubMed
-
- Stayner L., Steenland,K., Greife,A., Hornung,R., Hayes,R.B., Nowlin,S., Morawetz,J., Ringenburg,V., Elliot,L. and Halperin,W. (1993) Exposure-response analysis of cancer mortality in a cohort of workers exposed to ethylene oxide. Am. J. Epidemiol., 138, 787–798. - PubMed
-
- International Agency for Research on Cancer (IARC) (1985) IARC Monographs on the Evaluation of Carcinogenic Risk of Chemicals to Humans. Allyls Compounds, Aldehydes, Epoxides and Peroxides. Vol. 36. IARC, Lyon, France, pp. 189–226.

