Representational difference analysis in a lupus-prone mouse strain results in the identification of an unstable region of the genome on chromosome 11
- PMID: 11884638
- PMCID: PMC101347
- DOI: 10.1093/nar/30.6.1394
Representational difference analysis in a lupus-prone mouse strain results in the identification of an unstable region of the genome on chromosome 11
Abstract
BXSB mice develop a lupus-like autoimmune syndrome. We have previously identified several intervals that were linked to disease and, in an attempt to characterise lupus susceptibility genes within these intervals, we have sought to isolate differentially expressed genes. Representational difference analysis was used to compare gene expression between BXSB and C57BL/10 mice using spleen and thymus as a source of mRNA. The majority of differentially expressed sequences identified were immunoglobulin and gp70-related sequences, overexpression of these being characteristic of the disease. Among other isolated sequences were a sialyltransferase gene, a mouse tumour virus superantigen gene (Mtv-3), and the virus-related sequence, hitchhiker. In BXSB the sialyltransferase gene not only overexpressed spliced transcripts, but also produced high levels of unspliced mRNA. Further analysis demonstrated that the copy number of the three linked sequences: sialyltransferase, Mtv-3 and hitchhiker, was amplified in BXSB and that the structural organisation of this locus varies in different mouse strains. This locus consists of three parts, Mtv-3-hitchhiker-sialyltransferase, hitchhiker-sialyltransferase, and sialyltransferase alone. Different combinations of the regions are present in different mouse strains. Linkage analysis demonstrated that this region at the distal end of chromosome 11 is weakly linked to phenotypic markers of disease.
Figures

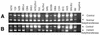

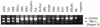

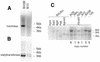
Similar articles
-
Identification of intervals on chromosomes 1, 3, and 13 linked to the development of lupus in BXSB mice.Arthritis Rheum. 2000 Feb;43(2):349-55. doi: 10.1002/1529-0131(200002)43:2<349::AID-ANR14>3.0.CO;2-M. Arthritis Rheum. 2000. PMID: 10693874
-
Multiple lupus susceptibility loci map to chromosome 1 in BXSB mice.J Immunol. 1998 Sep 15;161(6):2753-61. J Immunol. 1998. PMID: 9743333
-
The Y chromosome from autoimmune BXSB/MpJ mice induces a lupus-like syndrome in (NZW x C57BL/6)F1 male mice, but not in C57BL/6 male mice.Eur J Immunol. 1988 Jun;18(6):911-5. doi: 10.1002/eji.1830180612. Eur J Immunol. 1988. PMID: 3260184
-
Overlapping BXSB congenic intervals, in combination with microarray gene expression, reveal novel lupus candidate genes.Genes Immun. 2006 Apr;7(3):250-63. doi: 10.1038/sj.gene.6364294. Genes Immun. 2006. PMID: 16541099
-
Systemic lupus erythematosus--recent clues from congenic strains.Arch Immunol Ther Exp (Warsz). 2005 May-Jun;53(3):207-12. Arch Immunol Ther Exp (Warsz). 2005. PMID: 15995581 Review.
Cited by
-
APOBEC3 enzymes restrict marginal zone B cells.Eur J Immunol. 2015 Mar;45(3):695-704. doi: 10.1002/eji.201445218. Epub 2015 Jan 21. Eur J Immunol. 2015. PMID: 25501566 Free PMC article.
References
-
- Wakeland E.K., Wandstrat,A.E., Liu,K. and Morel,L. (1999) Genetic dissection of systemic lupus erythematosus. Curr. Opin. Immunol., 11, 701–707. - PubMed
-
- Vyse T.J. and Kotzin,B.L. (1998) Genetic susceptibility to systemic lupus erythematosus. Annu. Rev. Immunol., 16, 261–292. - PubMed
-
- Tsao B.P. (2000) Lupus susceptibility genes on human chromosome 1. Int. Rev. Immunol., 19, 319–334. - PubMed
-
- Kono D.H. and Theofilopoulos,A.N. (2000) Genetic studies in systemic autoimmunity and aging. Immunol. Res., 21, 111–122. - PubMed
-
- Yasutomo K., Horiuchi,T., Kagami,S., Tsukamoto,H., Hashimura,C., Urushihara,M. and Kuroda,Y. (2001) Mutation of DNASE1 in people with systemic lupus erythematosus. Nature Genet., 28, 313–314. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

