Characterization of phenylpropene O-methyltransferases from sweet basil: facile change of substrate specificity and convergent evolution within a plant O-methyltransferase family
- PMID: 11884690
- PMCID: PMC152928
- DOI: 10.1105/tpc.010327
Characterization of phenylpropene O-methyltransferases from sweet basil: facile change of substrate specificity and convergent evolution within a plant O-methyltransferase family
Abstract
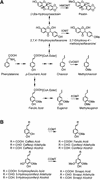
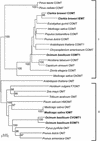
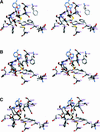
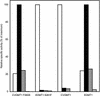
Some basil varieties are able to convert the phenylpropenes chavicol and eugenol to methylchavicol and methyleugenol, respectively. Chavicol O-methyltransferase (CVOMT) and eugenol O-methyltransferase (EOMT) cDNAs were isolated from the sweet basil variety EMX-1 using a biochemical genomics approach. These cDNAs encode proteins that are 90% identical to each other and very similar to several isoflavone O-methyltransferases such as IOMT, which catalyzes the 4'-O-methylation of 2,7,4'-trihydroxyisoflavanone. On the other hand, CVOMT1 and EOMT1 are related only distantly to (iso)eugenol OMT from Clarkia breweri, indicating that the eugenol O-methylating enzymes in basil and C. breweri evolved independently. Transcripts for CVOMT1 and EOMT1 were highly expressed in the peltate glandular trichomes on the surface of the young basil leaves. The CVOMT1 and EOMT1 cDNAs were expressed in Escherichia coli, and active proteins were produced. CVOMT1 catalyzed the O-methylation of chavicol, and EOMT1 also catalyzed the O-methylation of chavicol with equal efficiency to that of CVOMT1, but it was much more efficient in O-methylating eugenol. Molecular modeling, based on the crystal structure of IOMT, suggested that a single amino acid difference was responsible for the difference in substrate discrimination between CVOMT1 and EOMT1. This prediction was confirmed by site-directed mutagenesis, in which the appropriate mutants of CVOMT1 (F260S) and EOMT1 (S261F) were produced that exhibited the opposite substrate preference relative to the respective native enzyme.
Figures



References
-
- Adams, S., and Weidenborner, M. (1996). Mycelial deformations of Cladosporium herbarum due to the application of eugenol or carvacrol. J. Essential Oil Res. 8, 535–540.
-
- Bradford, M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. - PubMed
-
- Chenchik, A., Zhu, Y., Diatchenko, L., Li, R., Hill, J., and Siebert, P. (1996). Generation and use of high-quality cDNA from small amounts of total RNA by SMART PCR. In RT-PCR Methods for Gene Cloning and Analysis, P. Siebert and J. Larrick, eds (Westborough, MA: BioTechniques Books), pp. 305–319.
-
- Collendavelloo, J., Legrand, M., Geoffroy, P., Barthelemy, J., and Fritig, B. (1981). Purification and properties of the three o-diphenol-O-methyltransferases of tobacco leaves. Phytochemistry 20, 611–616.
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

