Arabidopsis FHY3 defines a key phytochrome A signaling component directly interacting with its homologous partner FAR1
- PMID: 11889039
- PMCID: PMC125923
- DOI: 10.1093/emboj/21.6.1339
Arabidopsis FHY3 defines a key phytochrome A signaling component directly interacting with its homologous partner FAR1
Abstract
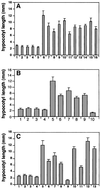
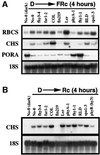
In Arabidopsis, phytochrome A (phyA) is the primary photoreceptor mediating various plant responses to far-red (FR) light. Here we show that phyA signaling involves a combinatorial action of downstream intermediates, which controls overlapping yet distinctive sets of FR responses. FHY3 is a prominent phyA signaling intermediate sharing structural similarity to FAR1, a previously identified phyA signaling component. The fhy3 and far1 mutants display similar yet distinctive defects in phyA signaling; however, overexpression of either FHY3 or FAR1 suppresses the mutant phenotype of both genes. Moreover, overexpression of partial fragments of FHY3 can cause a dominant-negative interference phenotype on phyA signaling that is stronger than those of the fhy3 or far1 null mutants. Further, we demonstrate that FHY3 and FAR1 are capable of homo- and hetero-interaction. Our data indicate that FHY3, together with FAR1, defines a key module in a signaling network underlying phyA-mediated FR light responses.
Figures





Similar articles
-
The FHY3 and FAR1 genes encode transposase-related proteins involved in regulation of gene expression by the phytochrome A-signaling pathway.Plant J. 2003 May;34(4):453-71. doi: 10.1046/j.1365-313x.2003.01741.x. Plant J. 2003. PMID: 12753585
-
Discrete and essential roles of the multiple domains of Arabidopsis FHY3 in mediating phytochrome A signal transduction.Plant Physiol. 2008 Oct;148(2):981-92. doi: 10.1104/pp.108.120436. Epub 2008 Aug 20. Plant Physiol. 2008. PMID: 18715961 Free PMC article.
-
Arabidopsis transcription factor ELONGATED HYPOCOTYL5 plays a role in the feedback regulation of phytochrome A signaling.Plant Cell. 2010 Nov;22(11):3634-49. doi: 10.1105/tpc.110.075788. Epub 2010 Nov 19. Plant Cell. 2010. PMID: 21097709 Free PMC article.
-
Emerging Roles of FHY3 and FAR1 as System Integrators in Plant Development.Plant Cell Physiol. 2023 Oct 16;64(10):1139-1145. doi: 10.1093/pcp/pcad068. Plant Cell Physiol. 2023. PMID: 37384577 Review.
-
Multifaceted roles of FHY3 and FAR1 in light signaling and beyond.Trends Plant Sci. 2015 Jul;20(7):453-61. doi: 10.1016/j.tplants.2015.04.003. Epub 2015 May 5. Trends Plant Sci. 2015. PMID: 25956482 Review.
Cited by
-
FAR-RED ELONGATED HYPOCOTYL3 activates SEPALLATA2 but inhibits CLAVATA3 to regulate meristem determinacy and maintenance in Arabidopsis.Proc Natl Acad Sci U S A. 2016 Aug 16;113(33):9375-80. doi: 10.1073/pnas.1602960113. Epub 2016 Jul 28. Proc Natl Acad Sci U S A. 2016. PMID: 27469166 Free PMC article.
-
Overexpression of a mutant basic helix-loop-helix protein HFR1, HFR1-deltaN105, activates a branch pathway of light signaling in Arabidopsis.Plant Physiol. 2003 Dec;133(4):1630-42. doi: 10.1104/pp.103.029751. Epub 2003 Nov 26. Plant Physiol. 2003. PMID: 14645731 Free PMC article.
-
Light regulates COP1-mediated degradation of HFR1, a transcription factor essential for light signaling in Arabidopsis.Plant Cell. 2005 Mar;17(3):804-21. doi: 10.1105/tpc.104.030205. Epub 2005 Feb 10. Plant Cell. 2005. PMID: 15705947 Free PMC article.
-
Arabidopsis FHY1 protein stability is regulated by light via phytochrome A and 26S proteasome.Plant Physiol. 2005 Nov;139(3):1234-43. doi: 10.1104/pp.105.067645. Epub 2005 Oct 21. Plant Physiol. 2005. PMID: 16244150 Free PMC article.
-
FHY3 and FAR1 Act Downstream of Light Stable Phytochromes.Front Plant Sci. 2016 Feb 22;7:175. doi: 10.3389/fpls.2016.00175. eCollection 2016. Front Plant Sci. 2016. PMID: 26941752 Free PMC article.
References
-
- Ausubel F., Brent,R., Kingston,R.E., Moore,D.D., Seidman,J.G., Smith, J.A. and Struhl,K. (1999) Short Protocols in Molecular Biology. John Wiley and Sons, New York, NY.
-
- Barnes S.A., Quaggio,R.B., Whitelam,G.C. and Chua,N.-H. (1996a) fhy1 defines a branch point in phytochrome A signal transduction pathways for gene expression. Plant J., 10, 1155–1161. - PubMed
-
- Bell C.J. and Ecker,J.R. (1994) Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics, 19, 137–144. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

