Isolation and characterization of cLV25, a Bacteroides fragilis chromosomal transfer factor resembling multiple Bacteroides sp. mobilizable transposons
- PMID: 11889096
- PMCID: PMC134936
- DOI: 10.1128/JB.184.7.1895-1904.2002
Isolation and characterization of cLV25, a Bacteroides fragilis chromosomal transfer factor resembling multiple Bacteroides sp. mobilizable transposons
Abstract
Horizontal DNA transfer contributes significantly to the dissemination of antibiotic resistance genes in Bacteroides fragilis. To further our understanding of DNA transfer in B. fragilis, we isolated and characterized a new transfer factor, cLV25. cLV25 was isolated from B. fragilis LV25 by its capture on the nonmobilizable Escherichia coli-Bacteroides shuttle vector pGAT400DeltaBglII. Similar to other Bacteroides sp. transfer factors, cLV25 was mobilized in E. coli by the conjugative plasmid R751. Using Tn1000 mutagenesis and deletion analysis of cLV25, two mobilization genes, bmgA and bmgB, were identified, whose predicted proteins have similarity to DNA relaxases and mobilization proteins, respectively. In particular, BmgA and BmgB were homologous to MocA and MocB, respectively, the two mobilization proteins of the B. fragilis mobilizable transposon Tn4399. A cis-acting origin of transfer (oriT) was localized to a 353-bp region that included nearly all of the intergenic region between bmgB and orf22 and overlapped with the 3' end of orf22. This oriT contained a putative nic site sequence but showed no significant similarity to the oriT regions of other transfer factors, including Tn4399. Despite the lack of sequence similarity between the oriTs of cLV25 and Tn4399, a mutation in the cLV25 putative DNA relaxase, bmgA, was partially complemented by Tn4399. In addition to the functional cross-reaction with Tn4399, a second distinguishing feature of cLV25 is that predicted proteins have similarity to proteins encoded not only by Tn4399 but by several Bacteroides sp. transfer factors, including NBU1, NBU2, CTnDOT, Tn4555, and Tn5520.
Figures


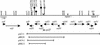

References
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1998. Current protocols in molecular biology. John Wiley and Sons, New York, N.Y.
-
- Bonheyo, G., D. Graham, N. B. Shoemaker, and A. A. Salyers. 2001. Transfer region of a Bacteroides conjugative transposon, CTnDOT. Plasmid 45:41-51. - PubMed
-
- Boyd, D. A., G. A. Peters, L. Ng, and M. R. Mulvey. 2000. Partial characterization of a genomic island associated with the multidrug resistance region of Salmonella enterica Typhimurium DT104. FEMS Microbiol. Lett. 189:285-291. - PubMed
-
- Chandler, M., and D. J. Galas. 1983. Cointegrate formation mediated by Tn9. II. Activity of IS1 is modulated by external DNA sequences. J. Mol. Biol. 170:61-91. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

