Two distinct alcohol dehydrogenases participate in butane metabolism by Pseudomonas butanovora
- PMID: 11889098
- PMCID: PMC134940
- DOI: 10.1128/JB.184.7.1916-1924.2002
Two distinct alcohol dehydrogenases participate in butane metabolism by Pseudomonas butanovora
Abstract
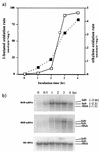
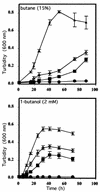
The involvement of two primary alcohol dehydrogenases, BDH and BOH, in butane utilization in Pseudomonas butanovora (ATCC 43655) was demonstrated. The genes coding for BOH and BDH were isolated and characterized. The deduced amino acid sequence of BOH suggests a 67-kDa alcohol dehydrogenase containing pyrroloquinoline quinone (PQQ) as cofactor and in the periplasm (29-residue leader sequence). The deduced amino acid sequence of BDH is consistent with a 70.9-kDa, soluble, periplasmic (37-residue leader sequence) alcohol dehydrogenase containing PQQ and heme c as cofactors. BOH and BDH mRNAs were induced whenever the cell's 1-butanol oxidation activity was induced. When induced with butane, the gene for BOH was expressed earlier than the gene for BDH. Insertional disruption of bdh or boh affected adversely, but did not eliminate, butane utilization by P. butanovora. The P. butanovora mutant with both genes boh and bdh inactivated was unable to grow on butane or 1-butanol. These cells, when grown in citrate and incubated in butane, developed butane oxidation capability and accumulated 1-butanol. The enzyme activity of BOH was characterized in cell extracts of the P. butanovora strain with bdh disrupted. Unlike BDH, BOH oxidized 2-butanol. The results support the involvement of two distinct NAD(+)-independent, PQQ-containing alcohol dehydrogenases, BOH (a quinoprotein) and BDH (a quinohemoprotein), in the butane oxidation pathway of P. butanovora.
Figures
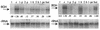

References
-
- Anzai, Y., H. Kim, J. Y. Park, H. Wakabayashi, and H. Oyaizu. 2000. Phylogenetic affiliation of the pseudomonads based on 16S rRNA sequence. Int. J. Syst. E vol. Microbiol. 50:1563-1589. - PubMed
-
- Arp, D. J. 1999. Butane metabolism by butane-grown Pseudomonas butanovora. Microbiology 145:1173-1180. - PubMed
-
- Ashraf, W., A. Mihdhir, and J. C. Murrell. 1994. Bacterial oxidation of propane. FEMS Microbiol. Lett. 122:1-6. - PubMed
-
- Ashraf, W., and J. C. Murrell. 1992. Genetic, biochemical and immunological evidence for the involvement of two alcohol dehydrogenases in the metabolism of propane by Rhodococcus rhodochrous PNKb1. Arch. Microbiol. 157:488-492.
-
- Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:2482-2488. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

