Identification and characterization of CAMP cohemolysin as a potential virulence factor of Riemerella anatipestifer
- PMID: 11889100
- PMCID: PMC134935
- DOI: 10.1128/JB.184.7.1932-1939.2002
Identification and characterization of CAMP cohemolysin as a potential virulence factor of Riemerella anatipestifer
Abstract
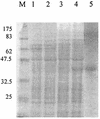
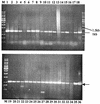
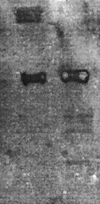
Riemerella anatipestifer is responsible for exudative septicemia in ducks. The genetic determinant of the CAMP cohemolysin, cam, from a strain of R. anatipestifer was cloned and expressed in Escherichia coli. Chromosomal DNA from serotype 19 strain 30/90 was used to construct a gene library in pBluescript II SK(-) vector in E. coli XL-1-Blue strain. The clones containing recombinant plasmids were screened for the CAMP reaction with Staphylococcus aureus. Those that showed cohemolysis were chosen for further analysis by sequencing. One of these clones, JFRA8, was subcloned to identify the smallest possible DNA fragment containing the CAMP cohemolysin determinant, which was located on a 3,566-bp BamHI-BstXI fragment which specified a 1,026-bp open reading frame. Clones containing recombinant plasmids carrying cam obtained by PCR cloning into E. coli M15 strain secreted an active CAMP cohemolysin. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blot analyses confirmed that the recombinant strain expressed a protein with a molecular mass of 37 kDa and that strains from serotypes 1, 2, 3, 5, 6, and 19 expressed the cohemolysin. The deduced amino acid sequence showed high homology to those of O-sialoglycoprotein endopeptidases. Hydrolysis of radioiodinated glycophorin A confirmed that Cam is a sialoglycoprotease.
Figures




Similar articles
-
Identification, cloning, and expression of the CAMP-like factor autotransporter gene (cfa) of Bartonella henselae.Infect Immun. 2005 Jul;73(7):4205-13. doi: 10.1128/IAI.73.7.4205-4213.2005. Infect Immun. 2005. PMID: 15972511 Free PMC article.
-
Cloning and expression of a cohemolysin, the CAMP factor of Actinobacillus pleuropneumoniae.Infect Immun. 1989 Jul;57(7):2050-6. doi: 10.1128/iai.57.7.2050-2056.1989. Infect Immun. 1989. PMID: 2659534 Free PMC article.
-
Identification, cloning, and expression of the CAMP factor gene (cfa) of group A streptococci.Infect Immun. 1999 Sep;67(9):4725-31. doi: 10.1128/IAI.67.9.4725-4731.1999. Infect Immun. 1999. PMID: 10456923 Free PMC article.
-
Cloning, sequencing and expression of the CAMP factor gene of Streptococcus uberis.Microb Pathog. 1996 May;20(5):297-307. doi: 10.1006/mpat.1996.0028. Microb Pathog. 1996. PMID: 9132527
-
Development of an ELISA using a recombinant 41 kDa partial protein (P45N') for the detection of Riemerella anatipestifer infections in ducks.Vet Microbiol. 2002 Sep 24;88(4):339-49. doi: 10.1016/s0378-1135(02)00123-2. Vet Microbiol. 2002. PMID: 12220809
Cited by
-
Cardiolipin synthetase is involved in antagonistic interaction (reverse CAMP phenomenon) of Mycoplasma species with Staphylococcus aureus beta-hemolysis.J Clin Microbiol. 2014 May;52(5):1622-8. doi: 10.1128/JCM.00037-14. Epub 2014 Mar 5. J Clin Microbiol. 2014. PMID: 24599982 Free PMC article.
-
The Role of the Regulator Fur in Gene Regulation and Virulence of Riemerella anatipestifer Assessed Using an Unmarked Gene Deletion System.Front Cell Infect Microbiol. 2017 Aug 25;7:382. doi: 10.3389/fcimb.2017.00382. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 28971067 Free PMC article.
-
A Novel RAYM_RS09735/RAYM_RS09740 Two-Component Signaling System Regulates Gene Expression and Virulence in Riemerella anatipestifer.Front Microbiol. 2017 Apr 21;8:688. doi: 10.3389/fmicb.2017.00688. eCollection 2017. Front Microbiol. 2017. PMID: 28484437 Free PMC article.
-
Genome-Wide Analysis and Characterization of the Riemerella anatipestifer Putative T9SS Secretory Proteins with a Conserved C-Terminal Domain.J Bacteriol. 2022 Jul 19;204(7):e0007322. doi: 10.1128/jb.00073-22. Epub 2022 Jun 7. J Bacteriol. 2022. PMID: 35670588 Free PMC article.
-
The Riemerella anatipestifer AS87_01735 Gene Encodes Nicotinamidase PncA, an Important Virulence Factor.Appl Environ Microbiol. 2016 Sep 16;82(19):5815-23. doi: 10.1128/AEM.01829-16. Print 2016 Oct 1. Appl Environ Microbiol. 2016. PMID: 27451449 Free PMC article.
References
-
- Brogden, K. A. 1989. Pasteurella infection, p. 115-129. In C. Adlam and J. M. Rutter (ed.), Pasteurella and pasteurellosis. Academic Press, London, United Kingdom.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

