Control of butanol formation in Clostridium acetobutylicum by transcriptional activation
- PMID: 11889105
- PMCID: PMC134926
- DOI: 10.1128/JB.184.7.1966-1973.2002
Control of butanol formation in Clostridium acetobutylicum by transcriptional activation
Abstract
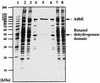
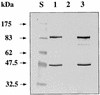
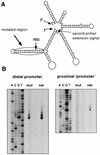
The sol operon of Clostridium acetobutylicum is the essential transcription unit for formation of the solvents butanol and acetone. The recent proposal that transcriptional regulation of this operon is controlled by the repressor Orf5/SolR (R. V. Nair, E. M. Green, D. E. Watson, G. N. Bennett, and E. T. Papoutsakis, J. Bacteriol. 181:319-330, 1999) was found to be incorrect. Instead, regulation depends on activation, most probably by the multivalent transcription factor Spo0A. The operon is transcribed from a single promoter. A second signal identified in primer extension studies results from mRNA processing and can be observed only in the natural host, not in a heterologous host. The first structural gene in the operon (adhE, encoding a bifunctional butyraldehyde/butanol dehydrogenase) is translated into two different proteins, the mature AdhE enzyme and the separate butanol dehydrogenase domain. The promoter of the sol operon is preceded by three imperfect repeats and a putative Spo0A-binding motif, which partially overlaps with repeat 3 (R3). Reporter gene analysis performed with the lacZ gene of Thermoanaerobacterium thermosulfurigenes and targeted mutations of the regulatory region revealed that the putative Spo0A-binding motif, R3, and R1 are essential for control. The data obtained also indicate that an additional activator protein is involved.
Figures



Similar articles
-
Regulation of the sol locus genes for butanol and acetone formation in Clostridium acetobutylicum ATCC 824 by a putative transcriptional repressor.J Bacteriol. 1999 Jan;181(1):319-30. doi: 10.1128/JB.181.1.319-330.1999. J Bacteriol. 1999. PMID: 9864345 Free PMC article.
-
Orf5/SolR: a transcriptional repressor of the sol operon of Clostridium acetobutylicum?J Ind Microbiol Biotechnol. 2001 Nov;27(5):307-13. doi: 10.1038/sj.jim.7000144. J Ind Microbiol Biotechnol. 2001. PMID: 11781806
-
Transcriptional regulation of solventogenesis in Clostridium acetobutylicum.J Mol Microbiol Biotechnol. 2002 May;4(3):295-300. J Mol Microbiol Biotechnol. 2002. PMID: 11931561 Review.
-
Molecular characterization and transcriptional analysis of adhE2, the gene encoding the NADH-dependent aldehyde/alcohol dehydrogenase responsible for butanol production in alcohologenic cultures of Clostridium acetobutylicum ATCC 824.J Bacteriol. 2002 Feb;184(3):821-30. doi: 10.1128/JB.184.3.821-830.2002. J Bacteriol. 2002. PMID: 11790753 Free PMC article.
-
Solventogenic enzymes of Clostridium acetobutylicum: catalytic properties, genetic organization, and transcriptional regulation.FEMS Microbiol Rev. 1995 Oct;17(3):251-62. doi: 10.1111/j.1574-6976.1995.tb00209.x. FEMS Microbiol Rev. 1995. PMID: 7576767 Review.
Cited by
-
A Quantitative System-Scale Characterization of the Metabolism of Clostridium acetobutylicum.mBio. 2015 Nov 24;6(6):e01808-15. doi: 10.1128/mBio.01808-15. mBio. 2015. PMID: 26604256 Free PMC article.
-
Small and Low but Potent: the Complex Regulatory Role of the Small RNA SolB in Solventogenesis in Clostridium acetobutylicum.Appl Environ Microbiol. 2018 Jul 2;84(14):e00597-18. doi: 10.1128/AEM.00597-18. Print 2018 Jul 15. Appl Environ Microbiol. 2018. PMID: 29728392 Free PMC article.
-
An improved kinetic model for the acetone-butanol-ethanol pathway of Clostridium acetobutylicum and model-based perturbation analysis.BMC Syst Biol. 2011 Jun 20;5 Suppl 1(Suppl 1):S12. doi: 10.1186/1752-0509-5-S1-S12. BMC Syst Biol. 2011. PMID: 21689471 Free PMC article.
-
Synthetic Biology Tools for Genome and Transcriptome Engineering of Solventogenic Clostridium.Front Bioeng Biotechnol. 2020 Apr 16;8:282. doi: 10.3389/fbioe.2020.00282. eCollection 2020. Front Bioeng Biotechnol. 2020. PMID: 32363182 Free PMC article. Review.
-
Genome-scale model for Clostridium acetobutylicum: Part I. Metabolic network resolution and analysis.Biotechnol Bioeng. 2008 Dec 1;101(5):1036-52. doi: 10.1002/bit.22010. Biotechnol Bioeng. 2008. PMID: 18767192 Free PMC article.
References
-
- Bertram, J., A. Kuhn, and P. Dürre. 1990. Tn916-induced mutants of Clostridium acetobutylicum defective in regulation of solvent formation. Arch. Microbiol. 153:373-377.
-
- Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. - PubMed
-
- Burchhardt, G., and H. Bahl. 1991. Cloning and analysis of the β-galactosidase-encoding gene from Clostridium thermosulfurogenes EM1. Gene 106:13-19. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

