Snakin-2, an antimicrobial peptide from potato whose gene is locally induced by wounding and responds to pathogen infection
- PMID: 11891250
- PMCID: PMC152207
- DOI: 10.1104/pp.010685
Snakin-2, an antimicrobial peptide from potato whose gene is locally induced by wounding and responds to pathogen infection
Abstract
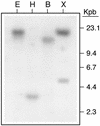
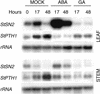
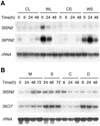
The peptide snakin-2 (StSN2) has been isolated from potato (Solanum tuberosum cv Jaerla) tubers and found to be active (EC(50) = 1-20 microM) against fungal and bacterial plant pathogens. It causes a rapid aggregation of both Gram-positive and Gram-negative bacteria. The corresponding StSN2 cDNA encodes a signal sequence followed by a 15-residue acidic sequence that precedes the mature StSN2 peptide, which is basic (isoelectric point = 9.16) and 66 amino acid residues long (molecular weight of 7,025). The StSN2 gene is developmentally expressed in tubers, stems, flowers, shoot apex, and leaves, but not in roots, or stolons, and is locally up-regulated by wounding and by abscisic acid treatment. Expression of this gene is also up-regulated after infection of potato tubers with the compatible fungus Botritys cinerea and down-regulated by the virulent bacteria Ralstonia solanacearum and Erwinia chrysanthemi. These observations are congruent with the hypothesis that the StSN2 is a component of both constitutive and inducible defense barriers.
Figures





References
-
- Agrios GN. Plant Pathology. Ed. 4. San Diego: Academic Press; 1997.
-
- Alamillo JM, García-Olmedo F. Effects of urate, a natural inhibitor of peroxynitrite-mediated toxicity, in the response of Arabidopsis thaliana to the bacterial pathogen Pseudomonas syringae. Plant J. 2001;25:529–541. - PubMed
-
- Aubert D, Chevillard M, Dorne AM, Arlaud G, Herzog M. Expression patterns of GASA genes in Arabidopsis thaliana: The GASA4 gene is up-regulated by gibberellins in meristematic regions. Plant Mol Biol. 1998;36:871–883. - PubMed
-
- Ben-Nissan G, Weiss D. The petunia homologue of tomato gast1: Transcript accumulation coincides with gibberellin-induced corolla cell elongation. Plant Mol Biol. 1996;32:1067–1074. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

