Embryonic stem cells and somatic cells differ in mutation frequency and type
- PMID: 11891338
- PMCID: PMC122567
- DOI: 10.1073/pnas.062527199
Embryonic stem cells and somatic cells differ in mutation frequency and type
Abstract
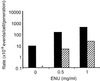
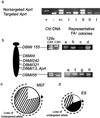
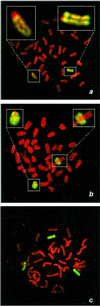
Pluripotent embryonic stem (ES) cells have been used to produce genetically modified mice as experimental models of human genetic diseases. Increasingly, human ES cells are being considered for their potential in the treatment of injury and disease. Here we have shown that mutation in murine ES cells, heterozygous at the selectable Aprt locus, differs from that in embryonic somatic cells. The mutation frequency in ES cells is significantly lower than that in mouse embryonic fibroblasts, which is similar to that in adult cells in vivo. The distribution of spontaneous mutagenic events is remarkably different between the two cell types. Although loss of the functional allele is the predominant mutation type in both cases, representing about 80% of all events, mitotic recombination accounted for all loss of heterozygosity events detected in somatic cells. In contrast, mitotic recombination in ES cells appeared to be suppressed and chromosome loss/reduplication, leading to uniparental disomy (UPD), represented more than half of the loss of heterozygosity events. Extended culture of ES cells led to accumulation of cells with adenine phosphoribosyltransferase deficiency and UPD. Because UPD leads to reduction to homozygosity at multiple recessive disease loci, including tumor suppressor loci, in the affected chromosome, the increased risk of tumor formation after stem cell therapy should be viewed with concern.
Figures
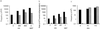

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

