Nuclear entry and CRM1-dependent nuclear export of the Rous sarcoma virus Gag polyprotein
- PMID: 11891341
- PMCID: PMC122628
- DOI: 10.1073/pnas.062652199
Nuclear entry and CRM1-dependent nuclear export of the Rous sarcoma virus Gag polyprotein
Abstract
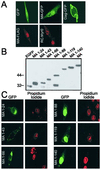
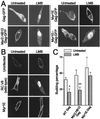
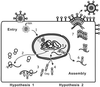
The retroviral Gag polyprotein directs budding from the plasma membrane of infected cells. Until now, it was believed that Gag proteins of type C retroviruses, including the prototypic oncoretrovirus Rous sarcoma virus, were synthesized on cytosolic ribosomes and targeted directly to the plasma membrane. Here we reveal a previously unknown step in the subcellular trafficking of the Gag protein, that of transient nuclear localization. We have identified a targeting signal within the N-terminal matrix domain that facilitates active nuclear import of the Gag polyprotein. We also found that Gag is transported out of the nucleus through the CRM1 nuclear export pathway, based on observations that treatment of virus-expressing cells with leptomycin B resulted in the redistribution of Gag proteins from the cytoplasm to the nucleus. Internal deletion of the C-terminal portion of the Gag p10 region resulted in the nuclear sequestration of Gag and markedly diminished budding, suggesting that the nuclear export signal might reside within p10. Finally, we observed that a previously described matrix mutant, Myr1E, was insensitive to the effects of leptomycin B, apparently bypassing the nuclear compartment during virus assembly. Myr1E has a defect in genomic RNA packaging, implying that nuclear localization of Gag might be involved in viral RNA interactions. Taken together, these findings provide evidence that nuclear entry and egress of the Gag polyprotein are intrinsic components of the Rous sarcoma virus assembly pathway.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

