Genome scans provide evidence for low-HDL-C loci on chromosomes 8q23, 16q24.1-24.2, and 20q13.11 in Finnish families
- PMID: 11891617
- PMCID: PMC447608
- DOI: 10.1086/339988
Genome scans provide evidence for low-HDL-C loci on chromosomes 8q23, 16q24.1-24.2, and 20q13.11 in Finnish families
Abstract
We performed a genomewide scan for genes that predispose to low serum HDL cholesterol (HDL-C) in 25 well-defined Finnish families that were ascertained for familial low HDL-C and premature coronary heart disease. The potential loci for low HDL-C that were identified initially were tested in an independent sample group of 29 Finnish families that were ascertained for familial combined hyperlipidemia (FCHL), expressing low HDL-C as one component trait. The data from the previous genome scan were also reanalyzed for this trait. We found evidence for linkage between the low-HDL-C trait and three loci, in a pooled data analysis of families with low HDL-C and FCHL. The strongest statistical evidence was obtained at a locus on chromosome 8q23, with a two-point LOD score of 4.7 under a recessive mode of inheritance and a multipoint LOD score of 3.3. Evidence for linkage also emerged for loci on chromosomes 16q24.1-24.2 and 20q13.11, the latter representing a recently characterized region for type 2 diabetes. Besides these three loci, loci on chromosomes 2p and 3p showed linkage in the families with low HDL-C and a locus on 2ptel in the families with FCHL.
Figures
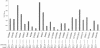
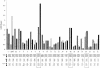
References
Electronic-Database Information
-
- Center for Medical Genetics, Marshfield Medical Research Foundation, http://research.marshfieldclinic.org/genetics/
-
- Cooperative Human Linkage Center, The, http://lpg.nci.nih.gov/CHLC/ (for map)
-
- Genetic Location Database (LDB), The, http://cedar.genetics.soton.ac.uk/public_html/ldb.html
-
- Genome Database, The, http://www.gdb.org/
References
-
- Bowden DW, Sale M, Howard TD, Qadri A, Spray BJ, Rothschild CB, Akots G, Rich SS, Freedman BI (1997) Linkage of genetic markers on human chromosomes 20 and 12 to NIDDM in Caucasian sib pairs with a history of diabetic nephropathy. Diabetes 46:882–886 - PubMed
-
- Brooks-Wilson A, Marcil M, Clee SM, Zhang LH, Roomp K, van Dam M, Yu L, Brewer C, Collins JA, Molhuizen HO, Loubser O, Ouelette BF, Fichter K, Ashbourne-Excoffon KJ, Sensen CW, Scherer S, Mott S, Denis M, Martindale D, Frohlich J, Morgan K, Koop B, Pimstone S, Kastelein JJ, Hayden MR (1999) Mutations in ABC1 in Tangier disease and familial high-density lipoprotein deficiency. Nat Genet 22:336–345 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

