PKHD1, the polycystic kidney and hepatic disease 1 gene, encodes a novel large protein containing multiple immunoglobulin-like plexin-transcription-factor domains and parallel beta-helix 1 repeats
- PMID: 11898128
- PMCID: PMC447605
- DOI: 10.1086/340448
PKHD1, the polycystic kidney and hepatic disease 1 gene, encodes a novel large protein containing multiple immunoglobulin-like plexin-transcription-factor domains and parallel beta-helix 1 repeats
Abstract
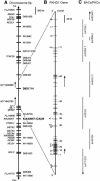
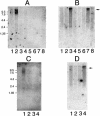
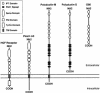
Autosomal recessive polycystic kidney disease (ARPKD) is a severe form of polycystic kidney disease that presents primarily in infancy and childhood and that is characterized by enlarged kidneys and congenital hepatic fibrosis. We have identified PKHD1, the gene mutated in ARPKD. PKHD1 extends over > or =469 kb, is primarily expressed in human fetal and adult kidney, and includes a minimum of 86 exons that are variably assembled into a number of alternatively spliced transcripts. The longest continuous open reading frame encodes a 4,074-amino-acid protein, polyductin, that is predicted to have a single transmembrane (TM)-spanning domain near its carboxyl terminus, immunoglobulin-like plexin-transcription-factor domains, and parallel beta-helix 1 repeats in its amino terminus. Several transcripts encode truncated products that lack the TM and that may be secreted if translated. The PKHD1-gene products are members of a novel class of proteins that share structural features with hepatocyte growth-factor receptor and plexins and that belong to a superfamily of proteins involved in regulation of cell proliferation and of cellular adhesion and repulsion.
Figures




Comment in
-
Autosomal recessive polycystic kidney disease (ARPKD): new insights from the identification of the ARPKD gene, PKHD1.Pediatr Res. 2002 Dec;52(6):830-1. doi: 10.1203/00006450-200212000-00002. Pediatr Res. 2002. PMID: 12438655 No abstract available.
References
Electronic-Database Information
-
- ExPASy Molecular Biology Server, http://ca.expasy.org/
-
- GenBank Overview, http://www.ncbi.nlm.nih.gov/Genbank/GenbankOverview.html (for sequences of all 86 exons and the composite cDNA with the longest ORF [accession number AF480064])
References
-
- Burge C, Karlin S (1997) Prediction of complete gene structures in human genomic DNA. J Mol Biol 268:78–94 - PubMed
-
- Burset M, Guigo R (1996) Evaluation of gene structure prediction programs. Genomics 34:353–367 - PubMed
-
- Calvet JP (1993) Polycystic kidney disease: primary extracellular matrix abnormality or defective cellular differentiation? Kidney Int 43:101–108 - PubMed
-
- Eggermann T, Nothen MM, Propping P, Schwanitz G (1993) Molecular diagnosis of trisomy 18 using DNA recovered from paraffin embedded tissues and possible implications for genetic counselling Ann Genet 36:214–216 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

