Molecular features of the copper binding sites in the octarepeat domain of the prion protein
- PMID: 11900542
- PMCID: PMC2905306
- DOI: 10.1021/bi011922x
Molecular features of the copper binding sites in the octarepeat domain of the prion protein
Abstract
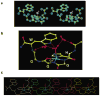
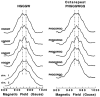
Recent evidence suggests that the prion protein (PrP) is a copper binding protein. The N-terminal region of human PrP contains four sequential copies of the highly conserved octarepeat sequence PHGGGWGQ spanning residues 60-91. This region selectively binds Cu2+ in vivo. In a previous study using peptide design, EPR, and CD spectroscopy, we showed that the HGGGW segment within each octarepeat comprises the fundamental Cu2+ binding unit [Aronoff-Spencer et al. (2000) Biochemistry 40, 13760-13771]. Here we present the first atomic resolution view of the copper binding site within an octarepeat. The crystal structure of HGGGW in a complex with Cu2+ reveals equatorial coordination by the histidine imidazole, two deprotonated glycine amides, and a glycine carbonyl, along with an axial water bridging to the Trp indole. Companion S-band EPR, X-band ESEEM, and HYSCORE experiments performed on a library of 15N-labeled peptides indicate that the structure of the copper binding site in HGGGW and PHGGGWGQ in solution is consistent with that of the crystal structure. Moreover, EPR performed on PrP(23-28, 57-91) and an 15N-labeled analogue demonstrates that the identified structure is maintained in the full PrP octarepeat domain. It has been shown that copper stimulates PrP endocytosis. The identified Gly-Cu linkage is unstable below pH approximately 6.5 and thus suggests a pH-dependent molecular mechanism by which PrP detects Cu2+ in the extracellular matrix or releases PrP-bound Cu2+ within the endosome. The structure also reveals an unusual complementary interaction between copper-structured HGGGW units that may facilitate molecular recognition between prion proteins, thereby suggesting a mechanism for transmembrane signaling and perhaps conversion to the pathogenic form.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

